Publications
2022: "Rationally designed immunogens enable immune focusing following SARS-CoV-2 spike imprinting," Blake M. Hauser, Maya Sangesland, Kerri J. St. Denis, Evan C. Lam, James Brett Case, Ian W. Windsor, Jared Feldman, Timothy M. Caradonna, Ty Kannegieter, Michael S. Diamond, Alejandro B. Balazs, Daniel Lingwood, Aaron G. Schmidt, Cell Rep. DOI: 10.1016/j.celrep.2022.110561 / NE-CAT 24-ID-E
2022: “Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN,” N.H. Moeller, K. Shi, O. Demir, C. Belica, S. Banerjee, L. Yin, C. Durfee, R.E. Amaro, H. Aihara, Proc. Natl. Acad. Sci. USA DOI: 10.1073/pnas.2106379119 / NE-CAT 24-ID-C
2022: "Structural assessment of HLA-A2-restricted SARS-CoV-2 spike epitopes recognized by public and private T-cell receptors," Daichao Wu, Alexander Kolesnikov, Rui Yin, Johnathan D. Guest, Ragul Gowthaman, Anton Shmelev, Yana Serdyuk, Dmitry V. Dianov, Grigory A. Efimov, Brian G. Pierce, Roy A. Mariuzza Nat. Commun. DOI: 10.1038/s41467-021-27669-8 / GM/CA-XSD 23-ID-D
2022: “Design, synthesis and in vitro evaluation of novel SARS-CoV-2 3CL(pro) covalent inhibitors,” J. K. Stille, J. Tjutrins, G. Wang, F. A. Venegas, C. Hennecker, A.M. Rueda, I. Sharon, N. Blaine, C. E. Miron, S. Pinus, A. Labarre, J. Plescia, M. Burai Patrascu, X. Zhang, A. S. Wahba, D. Vlaho, M. J. Huot, T. M. Schmeing, A. K. Mittermaier, N. Moitessier, Eur. J. Med. Chem. DOI: 10.1016/j.ejmech.2021.114046 / NE-CAT 24-ID-C
2022: “Crystal structure of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) frameshifting pseudoknot,” Christopher P. Jones and Adrian R. Ferre-D'Amare, RNA DOI: 10.1261/rna.078825.121 / NE-CAT 24-ID-C and 24-ID-E
2021: "Mechanisms of SARS-CoV-2 neutralization by shark variable new antigen receptors elucidated through X-ray crystallography," O. C. Ubah, E. W. Lake, G. S. Gunaratne, J. P. Gallant, M. Fernie, A. J. Robertson, J. S. Marchant, T. D. Bold, R. A. Langlois, W. E. Matchett, J. M. Thiede, K. Shi, L., Yin, N. H. Moeller, S. Banerjee, L. Ferguson, M. Kovaleva, A. J. Porter, H. Aihara, A. M. LeBeau, C. J. Barelle, Nat Commun. DOI: doi.org/10.1038/s41467-021-27611-y NE-CAT 24-ID-C
2021: “Low-dose in vivo protection and neutralization across SARS-CoV-2 variants by monoclonal antibody combinations,” V. Dussupt, R.S. Sankhala, L. Mendez-Rivera, S. M. Townsley, F. Schmidt, L. Wieczorek, K. G. Lal, G. C. Donofrio, U. Tran, N. D. Jackson, W. I. Zaky, M. Zemil, S. R. Tritsch, W. H. Chen, E. J. Martinez, A. Ahmed, M. Choe, W. C. Chang, A. Hajduczki, N. Jian, C. E. Peterson, P. A. Rees, M. Rutkowska, B. M. Slike, C. N. Selverian, I. Swafford, I. T. Teng, P. V, Thomas, T. Zhou, C. J. Smith, J. R. Currier, P. D. Kwong, M. Rolland, E. Davidson, B. J. Doranz, C. N. Mores, T. Hatziioannou, W. W, Reiley, P. D. Bieniasz, D. Paquin-Proulx, G. D. Gromowski, V. R. Polonis, N. L. Michael, K. Modjarrad, M. G. Joyce, S. J. Krebs, Nat. Immunol. DOI: 10.1038/s41590-021-01068-z / NE-CAT 24-ID-C & 24-ID-E, and SBC-XSD 19-BM
2021: "An Oral SARS-CoV-2 Mpro Inhibitor Clinical Candidate for the Treatment of COVID-19," Dafydd R. Owen, Charlotte M. N. Allerton, Annaliesa S. Anderson, Lisa Aschenbrenner, Melissa Avery, Simon Berritt, Britton Boras, Rhonda D. Cardin, Anthony Carlo, Karen J. Coffman, Alyssa Dantonio, Li Di, Heather Eng, RoseAnn Ferre, Ketan S. Gajiwala, Scott A. Gibson, Samantha E. Greasley, Brett L. Hurst, Eugene P. Kadar, Amit S. Kalgutkar, Jack C. Lee, Jisun Lee, Wei Liu, Stephen W. Mason, Stephen Noell, Jonathan J. Novak3, R. Scott Obach, Kevin Ogilvie, Nandini C. Patel, Martin Pettersson, Devendra K. Rai, Matthew R. Reese, Matthew F. Sammons, Jean G. Sathish, Ravi Shankar P. Singh, Claire M. Steppan, Al E. Stewart, Jamison B. Tuttle, Lawrence Updyke, Patrick R. Verhoest, Liuqing Wei, Qingyi Yang Yuao Zhu, Science 2 Nov 2021, DOI: 10.1126/science.abl4784 / IMCA-CAT 17-ID
2021: “A molecular sensor determines the ubiquitin substrate specificity of SARS-CoV-2 papain-like protease,” S. Patchett, Z. Lv, W. Rut, M. Bekes, M. Drag, S.K. Olsen, T.T. Huang, Cell Rep. DOI: 10.1016/j.celrep.2021.109754 / NE-CAT 24-ID-E
2021: "Structural Basis for SARS-CoV-2 Nucleocapsid Protein Recognition by Single-Domain Antibodies," Q. Ye, S. Lu, K.D. Corbett, Front. Immunol. DOI: 10.3389/fimmu.2021.719037 / NE-CAT 24-ID-E
2021: "Recognition of Divergent Viral Substrates by the SARS-CoV-2 Main Protease,” E.A. MacDonald, G. Frey, M.N. Namchuk, S.C. Harrison, S.M. Hinshaw, and I.W. Windsor, ACS Infect. Dis. DOI: 10.1021/acsinfecdis.1c00237 / NE-CAT 24-ID-C
2021: “Transient and stabilized complexes of Nsp7, Nsp8, and Nsp12 in SARS-CoV-2 replication,”Mateusz Wilamowski, Michal Hammel, Wellington Leite, Qiu Zhang, Youngchang Kim, Kevin L. Weiss, Robert Jedrzejczak, Daniel J. Rosenberg, Yichong Fan, Jacek Wower, Jan C. Bierma, Altaf H. Sarker, Susan E. Tsutakawa, Sai Venkatesh Pingali, Hugh M. O’Neill, Andrzej Joachimiak, Greg L. Hura, Biophys. J. DOI: 10.1016/j.bpj.2021.06.006 / SBC-XSD 19-ID
2021: "The SARS-CoV-2 Programmed −1 Ribosomal Frameshifting Element Crystal Structure Solved to 2.09 Å Using Chaperone-Assisted RNA Crystallography," Christina Roman, Anna Lewicka, Deepak Koirala, Nan-Sheng Li, Joseph A. Piccirilli, ACS Chem. Biol. DOI: 10.1021/acschembio.1c00324 / NE-CAT 24-ID-C/24-ID-E
2021: "Structure-Based Optimization of ML300-Derived, Noncovalent Inhibitors Targeting the Severe Acute Respiratory Syndrome Coronavirus 3CL Protease (SARS-CoV‑2 3CLpro)," Sang Hoon Han, Christopher M. Goins, Tarun Arya, Woo-Jin Shin, Joshua Maw, Alice Hooper, Dhiraj P. Sonawane, Matthew R. Porter, Breyanne E. Bannister, Rachel D. Crouch, A. Abigail Lindsey, Gabriella Lakatos, Steven R. Martinez, Joseph Alvarado, Wendell S. Akers, Nancy S. Wang, Jae U. Jung, Jonathan D. Macdonald, Shaun R. Stauffer, J Med. Chem. DOI: 10.1021/acs.jmedchem.1c00598 / LS-CAT 21-ID-F
2021: “The development of Nanosota-1 as anti-SARS-CoV-2 nanobody drug candidates,” G. Ye, J. Gallant, J. Zheng, C. Massey, K. Shi, W., Tai, A. Odle, M. Vickers, J. Shang, Y. Wan, L. Du, H. Aihara, S. Perlman, A. LeBeau, A., F. Li, Elife DOI: 10.7554/eLife.64815. / NE-CAT 24-ID-E
2021: "Postinfection treatment with a protease inhibitor increases survival of mice with a fatal SARS-CoV-2 infection," Chamandi S. Dampalla, Jian Zheng, Krishani Dinali Perera, Lok-Yin Roy Wong, David K. Meyerholz, Harry Nhat Nguyen, Maithri M. Kashipathy, Kevin P. Battaile, Scott Lovell, Yunjeong Kim, Stanley Perlman, William C. Groutas, and Kyeong-Ok Chang, Proceedings of the National Academy of Sciences of the United States of America. DOI: org/10.1073/pnas.2101555118 / IMCA-CAT 17-ID-B
2021: "Masitinib is a broad coronavirus 3CL inhibitor that blocks replication of SARS-CoV-2," Nir Drayman, Krysten A. Jones, Saara-Anne Azizi, Heather M. Froggatt, Kemin Tan, Natalia Ivanovna Maltseva, Siquan Chen, Vlad Nicolaescu, Steve Dvorkin, Kevin Furlong, Rahul S. Kathayat, Mason R. Firpo, Vincent Mastrodomenico, Emily A. Bruce, Madaline M. Schmidt, Robert Jedrzejczak, Miguel Á. Muñoz-Alía, Brooke Schuster, Vishnu Nair, Jason W. Botten, Christopher B. Brooke, Susan C. Baker, Bryan C. Mounce, Nicholas S. Heaton, Bryan C. Dickinson, Andrzej Jaochimiak, Glenn Randall, and Savaş Tay, Science. DOI: 10.1126/science.abg5827 / SBC-XSD 19-ID-D
2021: "Optimization of Triarylpyridinone Inhibitors of the Main Protease of SARS-CoV-2 to Low-Nanomolar Antiviral Potency," Chun-Hui Zhang, Krasimir A. Spasov, Raquel A. Reilly, Klarissa Hollander, Elizabeth A. Stone, Joseph A. Ippolito, Maria-Elena Liosi, Maya G. Deshmukh, Julian Tirado-Rives, Shuo Zhang, Zhuobin Liang, Scott J. Miller, Farren Isaacs, Brett D. Lindenbach, Karen S. Anderson, and William L. Jorgensen, ACS Medicinal Chemistry Letters. DOI: 10.1021/acsmedchemlett.1c00326 / NE-CAT 24-ID-E
2021: "Multivalency transforms SARS-CoV-2 antibodies into broad and ultrapotent neutralizers," Edurne Rujas, Iga Kucharska, Yong Zi Tan, Samir Benlekbir, Hong Cui, Tiantian Zhao, Gregory A. Wasney, Patrick Budylowski, Furkan Guvenc, Jocelyn C. Newton, Taylor Sicard, Anthony Semesi, Krithika Muthuraman, Amy Nouanesengsy, Katherine Prieto, Stephanie A. Bueler, Sawsan Youssef, Sindy Liao-Chan, Jacob Glanville, Natasha Christie-Holmes, Samira Mubareka, Scott D. Gray-Owen, John L. Rubinstein, Bebhinn Treanor, and Jean-Philippe Julien, Nature Communications. DOI: 10.1038/s41467-021-23825-2 / GM/CA-XSD 23-ID-D
2021: "Mn2+ coordinates Cap-0-RNA to align substrates for efficient 2′-O-methyl transfer by SARS-CoV-2 nsp16," George Minasov, Monica Rosas-Lemus, Ludmilla Shuvalova, Nicole L. Inniss, Joseph S. Brunzelle, Courtney M. Daczkowski, Paul Hoover, Andrew D. Mesecar, and Karla J. F. Satchell, Science Signaling. DOI: 10.1126/scisignal.abh2071 /689/eabh2071/ LS-CAT 21-ID
2021: "Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease," Deshmukh, M. G., Ippolito, J. A., Zhang, C. H., Stone, E. A., Reilly, R. A., Miller, S. J., Jorgensen, W. L., and Anderson, K. S. Structure. DOI: 10.1016/j.str.2021.06.002 NE-CAT 24-ID-E/24-ID-C
2021: "Interaction of 8-Anilinonaphthalene-1-Sulfonate with SARS-CoV-2 main protease and its application as a fluorescent probe for inhibitor identification," Peerapon Deetanya, Kowit Hengphasatporn, Patcharin Wilasluck, Yasuteru Shigeta, Thanyada Rungrotmongkol, Kittikhun Wangkanont, Computational and Structural Biotechnology Journal. DOI: 10.1016/j.csbj.2021.05.053 / LS-CAT 21-ID-F
2021: "A metal ion orients SARS-CoV-2 mRNA to ensure accurate 2′-O methylation of its first nucleotide," Thiruselvam Viswanathan, Anurag Misra, Siu-Hong Chan, Shan Qi, Nan Dai, Shailee Arya, Luis Martinez-Sobrido, Yogesh K. Gupta, Nature Communications. DOI: 10.1038/s41467-021-23594-y / NE-CAT-24-ID
2021: "2'-O methylation of RNA Cap in SARS-CoV-2 captured by serial crystallography", M. Wilamowski, D. A. Sherrell, G. Minasov, Y. Kim, L. Shuvalova, A. Lavens, R. Chard, N. Maltseva, R. Jedrzejczak, M. Rosas-Lemus, N. Saint, I.T. Foster, K. Michalska, K.J.F. Satchell, and A. Joachimiak, Proceedings of the National Academy of Sciences of the United States of America. DOI: 10.1073/pnas.2100170118 / SBC-XSD 19-ID-D
2021: "Structural and functional ramifications of antigenic drift in recent SARS-CoV-2 variants," Meng Yuan, Deli Huang, Chang-Chun D. Lee, Nicholas C. Wu, Abigail M. Jackson, Xueyong Zhu, Hejun Liu, Linghang Peng, Marit J. van Gils, Rogier W. Sanders, Dennis R. Burton, S. Momsen Reincke, Harald Prüss, Jakob Kreye, David Nemazee, Andrew B. Ward, Ian A. Wilson, Science. DOI: 10.1126/science.abh1139 / GM/CA-XSD 23-ID-B
2021: "A combination of cross-neutralizing antibodies synergizes to prevent SARS-CoV-2 and SARS-CoV pseudovirus infection," Hejun Liu, Meng Yuan, Deli Huang, Sandhya Bangaru, Chang-Chun D. Lee, Linghang Peng, Xueyong Zhu, David Nemazee, Marit J. van Gils, Rogier W. Sanders, Hans-Christian Kornau, S. Momsen Reincke, Harald Prüss, Jakob Kreye, Nicholas C. Wu, Andrew B. Ward, Ian A. Wilson, Cell Host & Microbe. DOI: 10.1016/j.chom.2021.04.005 / GM/CA-XSD 23-ID
2021: "Modular basis for potent SARS-CoV-2 neutralization by a prevalent VH1-2-derived antibody class," Micah Rapp, Yicheng Guo, Eswar R. Reddem, Jian Yu, Lihong Liu, Pengfei Wang, Gabriele Cerutti, Phinikoula Katsamba, Jude S. Bimela, Fabiana A. Bahna, Seetha M. Mannepalli, Baoshan Zhang, Peter D. Kwong, Yaoxing Huang, David D. Ho, Lawrence Shapiro, Zizhang Sheng, Cell Reports. DOI: 10.1016/j.celrep.2021.108950 / NE-CAT 24-ID-C
2021: "The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in non-human primates," Bryan E. Jones, Patricia L. Brown-Augsburger, Kizzmekia S. Corbett, Kathryn Westendorf, Julian Davies, Thomas P. Cujec, Christopher M. Wiethoff, Jamie L. Blackbourne, Beverly A. Heinz, Denisa Foster, Richard E. Higgs, Deepa Balasubramaniam, Lingshu Wang, Roza Bidshahri, Lucas Kraft, Yuri Hwang, Stefanie Žentelis, Kevin R. Jepson, Rodrigo Goya, Maia A. Smith, David W. Collins, Samuel J. Hinshaw, Sean A. Tycho, Davide Pellacani, Ping Xiang, Krithika Muthuraman, Solmaz Sobhanifar, Marissa H. Piper, Franz J. Triana, Jorg Hendle, Anna Pustilnik, Andrew C. Adams, Shawn J. Berens, Ralph S. Baric, David R. Martinez, Robert W. Cross, Thomas W. Geisbert, Viktoriya Borisevich, Olubukola Abiona, Hayley M. Belli, Maren de Vries, Adil Mohamed, Meike Dittmann, Marie Samanovic, Mark J. Mulligan, Jory A. Goldsmith, Ching-Lin Hsieh, Nicole V. Johnson, Daniel Wrapp, Jason S. McLellan, Bryan C. Barnhart, Barney S. Graham, John R. Mascola, Carl L. Hansen, and Ester Falconer, Science Translational Medicine. DOI: 10.1126/scitranslmed.abf1906 / LRL-CAT 31-ID
2021: “Lead compounds for the development of SARS-CoV-2 3CL protease inhibitors,” S. Iketani, F. Forouhar, H. Liu, S.J. Hong, F.Y. Lin, M.S. Nair, A. Zask, Y. Huang, L. Xing, B.R. Stockwell, A. Chavez, and D.D. Ho, Nature Communications. DOI: 10.1038/s41467-021-22362-2 / NE-CAT 24-ID-E
2021: "SARS-CoV-2 evolution in an immunocompromised host reveals shared neutralization escape mechanisms," Sarah A. Clark, Lars E. Clark, Junhua Pan, Adrian Coscia, Lindsay G.A. McKay, Sundaresh Shankar, Rebecca I. Johnson, Vesna Brusic, Manish C. Choudhary, James Regan, Jonathan Z. Li, Anthony Griffiths, Jonathan Abraham, Cell. DOI: 10.1016/j.cell.2021.03.027 / NE-CAT 24-ID-E
2021: “Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite,” Gabriele Cerutti, Yicheng Guo, Tongqing Zhou, Jason Gorman, Myungjin Lee, Micah Rapp, Eswar R. Reddem, Jian Yu, Fabiana Bahna, Jude Bimela, Yaoxing Huang, Phinikoula S. Katsamba, Lihong Liu, Manoj S. Nair, Reda Rawi, Adam S. Olia, Pengfei Wang, Baoshan Zhang, Gwo-Yu huang, David D. Ho, Zizhang Sheng, Peter D. Kwong, and Lawrence Shapiro, Cell Host Microbe. DOI: 10.1016/j.chom.2021.03.005 / NE-CAT 24-ID-C
2021: “Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors,” C.A. Brosey, J.H. Houl, P. Katsonis, L.P.F. Balapiti-Modarage, S. Bommagani, A. Arvai, D. Moiani, A. Bacolla, T. Link, L.S. Warden, O. Lichtarge, D.E. Jones, Z. Ahmed, and J.A. Tainer, Progress in Biophysics and Molecular Biology. DOI: 10.1016/j.pbiomolbio.2021.02.002 / NE-CAT 24-ID-C
2021: “Potent Noncovalent Inhibitors of the Main Protease of SARS-CoV-2 from Molecular Sculpting of the Drug Perampanel Guided by Free Energy Perturbation Calculations,” C.H. Zhang, E.A. Stone, M. Deshmukh, J.A. Ippolito, M.M. Ghahremanpour, J. Tirado-Rives, K.A. Spasov, S. Zhang, Y. Takeo, S.N. Kudalkar, Z. Liang, F. Isaacs, B. Lindenbach, S.J. Miller, K.S. Anderson, and W.L. Jorgensen, ACS Central Science. DOI: 10.1021/acscentsci.1c00039 / NE-CAT 24-ID-C/24-ID-E
2021: “Structural basis for differential recognition of phosphohistidine-containing peptides by 1-pHis and 3-pHis monoclonal antibodies,” Rajasree Kalagiri, Robyn L. Stanfield, Jill Meisenhelder, James J. La Clair, Stephen R. Fuhs, Ian A. Wilson, and Tony Hunter, Proceedings of the National Academy of Sciences of the United States of America. DOI: 10.1073/pnas.2010644118 / GMCA-XSD 23-ID-D
2021: "Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2", Youngchang Kim, Jacek Wower, Natalia Maltseva, Changsoo Chang, Robert Jedrzejczak, Mateusz Wilamowski, Soowon Kang, Vlad Nicolaescu, Glenn Randall, Karolina Michalska, and Andrzej Joachimiak, Communications Biology. DOI: 10.1038/s42003-021-01735-9 / SBC-XSD 19-ID
2021: “Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors,” Jerzy Osipiuk, Saara-Anne Azizi, Steve Dvorkin, Michael Endres, Robert Jedrzejczak, Krysten A. Jones, Soowon Kang, Rahul S. Kathayat, Youngchang Kim, Vladislav G. Lisnyak, Samantha L. Maki, Vlad Nicolaescu, Cooper A. Taylor, Christine Tesar, Yu-An Zhang, Zhiyao Zhou, Glenn Randall, Karolina Michalska, Scott A. Snyder, Bryan C. Dickinson, and Andrzej Joachimiak, Nature Communications. DOI: 10.1038/s41467-021-21060-3 / SBC-XSD 19-ID
2021: “Structural characterization of Nonstructural protein 1 from SARS-CoV-2,” Cameron Semper, Nobuhiko Watanabe, and Alexei Savchenko, iScience. DOI: 10.1016/j.isci.2020.101903 / SBC-XSD 19-ID
2021: “The SARS-CoV-2 conserved macrodomain is a mono-ADP-ribosylhydrolase,” Yousef M. O. Alhammad, Maithri M. Kashipathy, Anuradha Roy, Jean-Philippe Gagné, Peter McDonald, Philip Gao, Louis Nonfoux, Kevin P. Battaile, David K. Johnson, Erik D. Holmstrom, Guy G. Poirier, Scott Lovell, Anthony R. Fehr, Journal of Virology. DOI: 10.1128/JVI.019 / IMCA-CAT 17-ID
2021: “Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape,” Paul-Albert König, Hrishikesh Das, Hejun Liu, Beate M. Kümmerer, Florian N. Gohr, Lea-Marie Jenster, Yonas M. Tesfamariam, Lisa D.J. Schiffelers, Miki Uchima, Jennifer D. Wuerth, Karl Gatterdam, Natalia Ruetalo, Maria H. Christensen, Caroline I. Fandrey, Sabine Normann, Steffen Pritzl, Jan M. P. Tödtmann, Leo Hanke, Jannik Boos, Meng Yuan, Xueyong Zhu, Jonathan Leo Schmid-Burgk, Hiroki Kato, Michael Schindler, Ian A. Wilson, Matthias Geyer, Kerstin U. Ludwig, B. Martin Hällberg, Nicholas C. Wu and Florian I. Schmidt, Science. DOI: 10.1126/science.abe6230 / GM/CA-XSD 23-ID-B
2020: "The SARS-CoV-2 Conserved Macrodomain Is a Mono-ADP-Ribosylhydrolase," Yousef M. O. Alhammad, Maithri M. Kashipathy, Anuradha Roy, Jean-Philippe Gagné, Peter McDonald, Philip Gao, Louis Nonfouxd, Kevin P. Battaile, David K. Johnson, Erik D. Holmstrom, Guy G. Poirier, Scott Lovell, Anthony R. Fehr, Journal of Virology, DOI: 10.1128/JVI.01969-20 / IMCA-CAT 17-ID
2020: "Structure and inhibition of the SARS-CoV-2 main protease reveals strategy for developing dual inhibitors against Mpro and cathepsin L", Michael Dominic Sacco, Chunlong Ma, Panagiotis Lagarias, Ang Gao, Julia Alma Townsend, Xiangzhi Meng, Peter Dube, Xiujun Zhang, Yanmei Hu, Naoya Kitamura, Brett Hurst, Bart Tarbet, Michael Thomas Marty, Antonios Kolocouris, Yan Xiang, Yu Chen, Jun Wang, Science Advances. DOI: 10.1126/sciadv.abe0751 / SBC-XSD 19-ID-D
2020: “Crystallographic structure of wild-type SARS-CoV-2 main protease acyl-enzyme intermediate with physiological C-terminal autoprocessing site,” Jaeyong Lee, Liam J. Worrall, Marija Vuckovic, Federico I. Rosell, Francesco Gentile, Anh-Tien Ton, Nathanael A. Caveney, Fuqiang Ban, Artem Cherkasov, Mark Paetzel, and Natalie C. J. Strynadka, Nature Communications. DOI: 10.1038/s41467-020-19662-4 / GM/CA-XSD 23-ID-B.
2020: "Versatile, Multivalent Nanobody Cocktails Efficiently Neutralize SARS-CoV-2", Yufei Xiang, Sham Nambulli, Zhengyun Xiao, Heng Liu, Zhe Sang, W. Paul Duprex, Dina Schneidman-Duhovny, Cheng Zhang, and Yi Shi, Science. DOI: 10.1126/science.abe4747 / GMCA-XSD 23-ID-B
2020: “Molecular determinants of vascular transport of dexamethasone in COVID-19 therapy,” Ivan G. Shabalin, Mateusz P. Czub, Karolina A. Majorek, Dariusz Brzezinski, Marek Grabowski, David R. Cooper, Mateusz Panasiuk, Maksymilian Chruszcz, and Wladek Minor, IUCrJ. DOI: 10.1107/S2052252520012944 / LS- CAT 21-ID-F
2020: "Structural basis for potent neutralization of SARS-CoV-2 and role of antibody affinity maturation," Nicholas K. Hurlburt, Emilie Seydoux, Yu-Hsin Wan, Venkata Viswanadh Edara, Andrew B. Stuart, Junli Feng, Mehul S. Suthar, Andrew T. McGuire, Leonidas Stamatatos, and Marie Pancera, Nature. Communications. DOI: 038/s41467-020-19231-9 / SBC-XSD 19-ID
2020: “Structure of human steroid 5α-reductase 2 with anti-androgen drug finasteride” by Qingpin Xiao, Lei Wang, Shreyas Supekar, Tao Shen, Heng Liu, Fei Ye, Junzhou Huang, Hao Fan, Zhiyi Wei, and Cheng Zhang, Nature Communications. DOI: 10.21203/rs.3.rs-40159/v1 / GM/CA-XSD 23-ID-B
2020: "Activity profiling and crystal structures of inhibitorbound SARS-CoV-2 papain-like protease: A framework for anti–COVID-19 drug design," Wioletta Rut, Zongyang Lv, Mikolaj Zmudzinski, Stephanie Patchett, Digant Nayak, Scott J. Snipas, Farid El Oualid, Tony T. Huang, Miklos Bekes, Marcin Drag, and Shaun K. Olsen, Science Advances. DOI: 10.1126/sciadv.abd4596 / NE-CAT 24-ID-C
2020: “Discovery of Ketone-Based Covalent Inhibitors of Coronavirus3CL Proteases for the Potential Therapeutic Treatment of COVID-19,” Robert L. Hoffman, Robert S. Kania, Mary A. Brothers, Jay F. Davies, Rose A. Ferre, Ketan S. Gajiwala, Mingying He, Robert J. Hogan, Kirk Kozminski, Lilian Y. Li, Jonathan W. Lockner, Jihong Lou, Michelle T. Marra, Lennert J. Mitchell Jr., Brion W. Murray, James A. Nieman, Stephen Noell, Simon P. Planken, Thomas Rowe, Kevin Ryan, George J. Smith III, James E. Solowiej, Claire M. Steppan, Barbara Taggart, Journal of Medicinal Chemistry. DOI: 10.1021/acs.jmedchem.0c01063 / IMCA-CAT 17-ID-B
2020: “High-resolution structures of the SARS-CoV-2 2′-O-methyltransferase reveal strategies for structure-based inhibitor design,” Monica Rosas-Lemus, George Minasov, Ludmilla Shuvalova, Nicole L. Inniss, Olga Kiryukhina, Joseph Brunzelle, and Karla J. F. Satchell, Science Signaling. DOI: 10.1126/scisignal.abe1202 / LS-CAT 21-ID-F
2020: “Discovery of Drug-like Ligands for the Mac1 Domain of SARS-CoV-2 Nsp3,” Rajdeep S. Virdi, Robert V. Bavisotto, Nicholas C. Hopper, Nemanja Vuksanovic, Trevor R. Melkonian, Nicholas R. Silvaggi, and David N. Frick SLAS Discovery. DOI: 10.1177/2472555220960428 / LS-CAT 21-ID-F
2020: “3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV–infected mice,” Athri D. Rathnayake, Jian Zheng, Yunjeong Kim, Krishani Dinali Perera3, Samantha Mackin, David K. Meyerholz, Maithri M. Kashipathy, Kevin P. Battaile, Scott Lovell, Stanley Perlman, William C. Groutas, and Kyeong-Ok Chang, Science Translational Medicine. DOI: 10.1126/scitranslmed.abc5332 / IMCA-CAT 17-ID-B
2020: “The Enzyme-Free Release of Nucleotides from Phosphoramidates Depends Strongly on the Amino Acid,” Dejana Jovanovic,Peter Tremmel, Pradeep S. Pallan, Martin Egli,* and Clemens Richert, Angewandte Chemie International Edition. DOI: 10.1002/anie.202008665 / LS-CAT 21-ID-F/21-ID-G
2020: “Structural basis of RNA cap modification by SARS-CoV-2,” Thiruselvam Viswanathan, Shailee Arya, Siu-Hong Chan, Shan Qi, Nan Dai, Anurag Misra, Jun-Gyu Park, Fatai Oladunni, Dmytro Kovalskyy, Robert A. Hromas, Luis Martinez-Sobrido, and Yogesh K. Gupta, Nature Communications. DOI: 10.1038/s41467-020-17496-8 / NE-CAT 24-ID-C
2020: “Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes,” Karolina Michalska, Youngchang Kim, Robert Jedrzejczak, Natalia I. Maltseva, Lucy Stols, Michael Endres, and Andrzej Joachimiak, IUCrJ. DOI: 10.1107/S2052252520009653 / SBC-XSD 9-ID-D
2020: “Architecture and self-assembly of the SARS-CoV-2 nucleocapsid protein,” Qiaozhen Ye, Alan M. V. West, Steve Silletti, and Kevin D Corbett, Protein Science. DOI: 10.1002/pro.3909 / NE-CAT 24-ID‐E
2020: “Molecular Basis for ADP-Ribose Binding to the Mac1 Domain of SARS-CoV-2 nsp3,” David N. Frick, Rajdeep S. Virdi, Nemanja Vuksanovic, Narayan Dahal, and Nicholas R. Silvaggi, Biochemistry. DOI: 10.1021/acs.biochem.0c00309 / LS-CAT 21-ID-F
2020: “Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease,” Chunlong Ma, Michael Dominic Sacco, Brett Hurst, Julia Alma Townsend, Yanmei Hu, Tommy Szeto, Xiujun Zhang, Bart Tarbet, Michael Thomas Marty, Yu Chen, and Jun Wang, Cell Research. DOI: 10.1038/s41422-020-0356-z / SBC-XSD 19-ID-D
2020: “A highly conserved cryptic epitope in the receptor-binding domains of SARS-CoV-2 and SARS-CoV,” Meng Yuan, Nicholas C. Wu, Xueyong Zhu, Chang-Chun D. Lee, Ray T. Y. So, Huibin Lv, Chris K. P. Mok, and Ian A. Wilson, Science. DOI: 10.1126/science.abb7269 / GM/CA-XSD 23-ID-D
2020: “Structural Basis for Potent Neutralization of Betacoronaviruses by Single-Domain Camelid Antibodies,” Daniel Wrapp, Dorien De Vlieger, Kizzmekia S. Corbett, Gretel M. Torres, Nianshuang Wang, Wander Van Breedam, Kenny Roose, Loes van Schie, VIB-CMB COVID-19 Response Team, Markus Hoffmann, Stefan Pöhlmann, Barney S. Graham, Nico Callewaert, Bert Schepens, Xavier Saelens, and Jason S. McLellan, Cell. DOI: 10.1016/j.cell.2020.04.031 / SBC-XSD 19-ID-D
2020: “Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2,” Youngchang Kim, Robert Jedrzejczak, Natalia I. Maltseva, Mateusz Wilamowski, Michael Endres, Adam Godzik, Karolina Michalska, and Andrzej Joachimiak, Protein Science. DOI: 10.1002/pro.3873 / SBC-XSD 19‐ID-D
2020: “Structural basis of receptor recognition by SARS-CoV-2,” Jian Shang, Gang Ye, Ke Shi, Yushun Wan, Chuming Luo, Hideki Aihara, Qibin Geng, Ashley Auerbach, and Fang Li, Nature. DOI: 10.1038/s41586-020-2179-y / NE-CAT 24-ID / The March 30, 2020, issue of the Guardian reported on the work.
Pre-Review Manuscripts
2022: Sophie M-C. Gobeil, Rory Henderson, Victoria Stalls, Katarzyna Janowska, Xiao Huang, Aaron May, Micah Speakman, Esther Beaudoin, Kartik Manne, Dapeng Li, Rob Parks, Maggie Barr, Margaret Deyton, Mitchell Martin, Katayoun Mansouri, Robert J. Edwards, Gregory D. Sempowski, Kevin O. Saunders, Kevin Wiehe, Wilton Williams, Bette Korber, Barton F. Haynes, Priyamvada Acharya, "Structural diversity of the SARS-CoV-2 Omicron spike," bioRxiv, DOI: 10.1101/2022.01.25.477784v1 / SER-CAT 22-ID
2021: J. Osipiuk, P.M. Wydorski, B.T. Lanham, C. Tesar, M. Endres, E. Engle, R. Jedrzejczak, V. Mullapudi, K. Michalska, K. Fidelis, D. Fushman, A. Joachimiak, L.A. Joachimiak, “Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin” bioRxiv, DOI: 10.1101/2021.09.15.460543 / SBC-XSD 19‐ID-D
2021: "LY-CoV1404 potently neutralizes SARS-CoV-2 variants," Kathryn Westendorf, Stefanie Žentelis, Denisa Foster, Peter Vaillancourt, Matthew Wiggin, Erica Lovett, Jörg Hendle, Anna Pustilnik, J. Michael Sauder, Lucas Kraft, Yuri Hwang, Robert W. Siegel, Jinbiao Chen, Beverly A. Heinz, Richard E. Higgs, Nicole Kalleward, Kevin Jepson, Rodrigo Goya, Maia A. Smith, David W. Collins, Davide Pellacani, Ping Xiang, Valentine de Puyraimond, Marketa Ricicova, Lindsay Devorkin, Caitlin Pritchard, Aoise O’Neill, Courtney Cohen, John Dye, Kathleen I. Huie, Catherine V. Badger, Darwyn Kobasa, Jonathan Audet, Joshua J. Freitas, Saleema Hassanali, Ina Hughes, Luis Munoz, Holly C. Palma, Bharathi Ramamurthy, Robert W. Cross, Thomas W. Geisbert, Viktoriya Borisevich, Iliana Lanz, Lisa Anderson, Payal Sipahimalani, Kizzmekia S. Corbett, Lingshu Wang, Eun Sung Yang, Yi Zhang, Wei Shi, Barney S. Graham, John R. Mascola, Tara L. Fernandez, Carl L. Hansen, Ester Falconer, Bryan E. Jones, and Bryan C. Barnhart, bioRxiv, DOI: 10.1101/2021.04.30.442182 / LRL-CAT 31-ID
2021: “Vitamin C inhibits SARS coronavirus-2 main protease essential for viral replication,” Tek Narsingh, Suraj Pandey, Ishwor Poudyal, Luis Aldama, Dennis Feliz, Moraima Noda, George N. Phillips Jr., Emina A. Stojković, and Marius Schmidt, bioRxiv. DOI: 10.1101/2021.05.02.442358 / SBC-XSD 19-ID-D
2021: “Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN,” N.H. Moeller, K. Shi, O. Demir, S. Banerjee, L. Yin, C. Belica, C. Durfee, R.E. Amaro, and H. Aihara, (2021) bioRxiv. DOI: 10.1101/2021.04.02.438274 NE-CAT 24-ID-C
2021: "Isolation and Characterization of Cross-Neutralizing Coronavirus Antibodies from COVID-19+ Subjects," Madelein Jennewein, Anna MacCamey, Nicholas Akins, Junli Feng, Leah Homad, Nicholas Hurlburt, Emily Seydoux, Yu-Hsin Wang, Andrew B Stuart, Venkata Vishwanadh Edara, Katharine Floyd, Abigail Vanderheiden, John R. Mascola, Nicole Doria-Rose, Lingshu Wang, Eun Yang, Helen Chu, Jonathan Torres, Gabriel Ozorowski, Andrew Ward, Rachael Whaley, Kristen Cohen, Marie Pancera, Juliana McElrath, Janet A Englund, Andres Finzi, Mehul Suthar, Andrew McGuire and Leonidas Stamatatos, bioRxiv. https://doi.org/10.1101/2021.03.23.436684 / SBC-XSD 19-ID
2021: "Inhibition of amyloid formation of the Nucleoprotein of SARS-CoV-2," Einav Tayeb-Fligelman, Xinyi Cheng, Christen Tai, Jeannette T. Bowler, Sarah Griner, Michael R. Sawaya, Paul M. Seidler, Yi Xiao Jiang, Jiahui Lu, Gregory M. Rosenberg, Lukasz Salwinski, Romany Abskharon, Chih-Te Zee, Ke Hou, Yan Li, David R. Boyer, Kevin A. Murray, Genesis Falcon, Daniel H. Anderson, Duilio Cascio, Lorena Saelices, Robert Damoiseaux, Feng Guo, David S. Eisenberg, bioRxiv. https://doi.org/10.1101/2021.03.05.434000 / NE-CAT 24-ID-C
2021: "Potent, Novel SARS-CoV-2 PLpro Inhibitors Block Viral Replication in Monkey and Human Cell Cultures," Zhengnan Shen, Kiira Ratia, Laura Cooper, Deyu Kong, Hyun Lee, Youngjin Kwon, Yangfeng Li, Saad Alqarni, Fei Huang, Oleksii Dubrovskyi, Lijun Rong, Gregory Thatcher, Rui Xiong, bioRxiv. https://doi.org/10.1101/2021.02.13.431008 / LS-CAT 21-ID-D,F,G
2021: "Synthetic nanobody–SARS-CoV-2 receptor-binding domain structures identify distinct epitopes," Javeed Ahmad, Jiansheng Jiang, Lisa F. Boyd, Kannan Natarajan, David H. Margulies, bioRxiv. DOI: 10.1101/2021.01.27.428466 / SER-CAT 22-ID, and BM
2020: “The Development of a Novel Nanobody Therapeutic for SARS-CoV-2,” Gang Ye, Joseph P. Gallant, Christopher Massey, Ke Shi, Wanbo Tai, Jian Zheng, Abby E. Odle, Molly A. Vickers, Jian Shang, Yushun Wan, Aleksandra Drelich, Kempaiah R. Kempaiah, Vivian Tat, Stanley Perlman, Lanying Du, Chien-Te Tseng, Hideki Aihara, Aaron M. LeBeau, Fang Li, bioRxiv. DOI: 10.1101/2020.11.17.386532v1 / NE-CAT 24-ID
2020: "Potent SARS-CoV-2 neutralizing antibodies selected from a human antibody library constructed decades ago", Min Qiang, Peixiang Ma, Yu Li, Hejun Liu, Adam Harding, Chenyu Min, Lili Liu, Meng Yuan, Qun Ji, Pingdong Tao, Xiaojie Shi, Zhean Li, Fulian Wang, Yu Zhang, Nicholas C. Wu, Chang-Chun D. Lee, Xueyong Zhu, Javier Gilbert-Jaramillo, Abhishek Saxena, Xingxu Huang, Hou Wang, William James, Raymond A. Dwek, Ian A. Wilson, Guang Yang, and Richard A. Lerner, bioRxiv. DOI: 10.1101/2020.11.06.370676 / GMCA-XSD 23-ID-B
2020: "Ebselen Reacts with SARS Coronavirus-1 2 Main Protease Crystals," Tek Narsingh Malla, Suraj Pandey, Ishwor Poudyal, Denisse Feliz, Moraima Noda, George Phillips, Emina Stojkovic, Marius Schmidt, bioRxiv. https://doi.org/10.1101/2020.08.10.244525 / SBC-XSD 19-ID-D
2020: "The crystal structure of nsp10-nsp16 heterodimer from SARS-CoV-2 in complex with S-adenosylmethionine", Monica Rosas-Lemus, George Minasov, Ludmilla Shuvalova, Nicole L. Inniss, Olga Kiryukhina, Grant Wiersum, Youngchang Kim, Robert Jedrzejczak, Natalia I. Maltseva, Michael Endres, Lukasz Jaroszewski, Adam Godzik, Andrzej Joachimiak, and Karla J. F. Satchell, bioRxiv. DOI: 10.1101/2020.04.17.047498 / SBC-XSD 19-ID-D
2020: "A Cryptic Site of Vulnerability on the Receptor Binding Domain of the SARS-CoV-2 Spike Glycoprotein", M. Gordon Joyce, Rajeshwer S. Sankhala, Wei-Hung Chen, Misook Choe, Hongjun Bai, Agnes Hajduczki, Lianying Yan, Spencer L. Sterling, Caroline E. Peterson, Ethan C. Green, Clayton Smith, Natalia de Val, Mihret Amare, Paul Scott, Eric D. Laing, Christopher C. Broder, Morgane Rolland, Nelson L. Michael, and Kayvon Modjarrad, bioRxiv. DOI: 10.1101/2020.03.15.992883 / NE-CAT 24-ID-C and NE-CAT 24-ID-E
SARS CoV-2 Research Highlights on the APS Web Site
"New, Potent Inhibitors of an Enzyme Essential for SARS-Cov-2 Viral Replication" 4.16.21
"The Structure of a Key Viral Enzyme Helps Identify Novel Drugs for Treatment of COVID-19" 3.1.21
"Discoveries from First SARS Outbreak Jump-Start COVID-19 Treatment Development" 1.25.21
"New Promising Antibodies against SARS-CoV-2" 1.15.21
"Deciphering Coronavirus Protein Behavior" 12.1.20
"Dexamethasone and COVID-19: All Patients Are Not Alike" 11.2.20
"Mapping the Structure of a Potent Antibody against the Coronavirus" 10.28.2020
"Viral 'Molecular Scissor' is Next COVID-19 Drug Target" 10.27.2020
"How SARS-CoV-2 RNA Evades Host Immune Responses" 08.26.2020
"3-D Structure of SARS-CoV-2 Explains High Infectivity vs. Other Coronaviruses" 06.10.2020
"New Coronavirus Protein Reveals Drug Target" 03.02.2020
SARS CoV-2 Feature Articles on the Argonne Web Site
Caught in the act: New data about COVID-19 virus’s functions could aid in treatment designs
Why the lovable llama might be a secret weapon against COVID-19
APS plays foundational role in development of COVID-19 vaccines
100th structure of COVID-19 virus from Advanced Photon Source data released
Hide and seek: Understanding how COVID-19 evades detection in a human cell
World’s X-ray facilities team up to battle COVID-19
10 ways Argonne science is combatting COVID-19
Eight ways Argonne advanced science in 2020
Advanced Photon Source at the heart of COVID-19 research
How scientists around the country are using the APS to fight COVID-19
Argonne’s researchers and facilities playing a key role in the fight against COVID-19
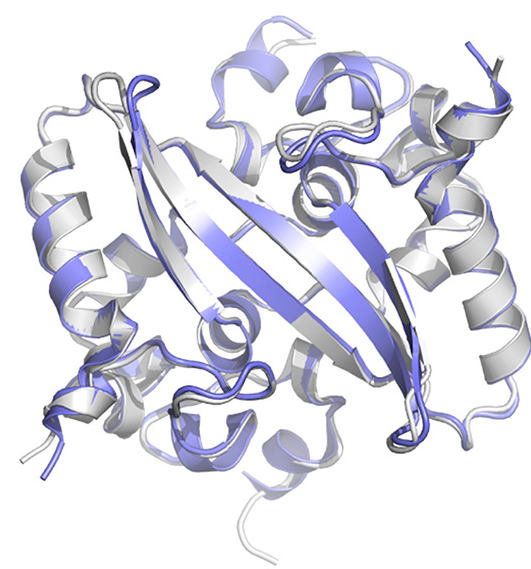 Structural overlay of the SARS‐CoV‐2 N2b dimer (blue) and the equivalent domain of SARS‐CoV‐N (PDB ID 2CJR). From Q. Ye et al., Protein Science. DOI: 10.1002/pro.3909 © 2020 The Protein Society / NE-CAT beamline 24-ID‐E
Structural overlay of the SARS‐CoV‐2 N2b dimer (blue) and the equivalent domain of SARS‐CoV‐N (PDB ID 2CJR). From Q. Ye et al., Protein Science. DOI: 10.1002/pro.3909 © 2020 The Protein Society / NE-CAT beamline 24-ID‐E
