Humans have been using enzymes to create new products for thousands of years. First it was wine, then cheese. In this tradition, three years ago, a team of scientists tweaked a lyase (HACL/S) to reverse course. Instead of breaking, the enzyme synthesizes novel chemicals through the addition of carbon atoms.
Now, using the Advanced Photon Source (APS), a U.S. Department of Energy (DOE) Office of Science user facility at DOE’s Argonne National Laboratory, an international team shows how HACL/S enzymes work on an atomic level. Their findings can serve as the basis for increasing the enzymes’ yield and versatility while drawing down as precursors atmospheric carbon dioxide and methane.
HACL/S enzymes were originally discovered for their role in breaking down fatty acids into formyl-CoA (fCoA) and an aldehyde or ketone in mammalian peroxisomes. Since then, scientists have discovered their ability to condense fCoA with various aldehydes and ketones and have one carbon atom added to them. Given the enzyme’s ability to reverse reaction direction from a lyase to a synthase, combined with an abundance of carbon molecules in the atmosphere, HACL/S is an ideal model for biocatalytic production of a variety of new products.
However, compared to chemical synthetic reactions, biocatalytic production usually produces low yield. The authors of the current research reasoned that if they could manipulate the specificity of these enzymes to accept different kinds of ketones or aldehydes, they could boost the enzymes’ productivity and efficiency.
In order to do that, they first needed to discover how these enzymes worked.
To begin, the team chose from the list of over 100 newly identified proteins six variants of the enzyme that exhibited high activity with aldehyde compounds of different length and formyl-CoA and had amino acid sequences that were diverse enough to cover the HACL/S subfamily. The team synthesized genes for each of the variants, then expressed them in Escherichia coli bacteria.
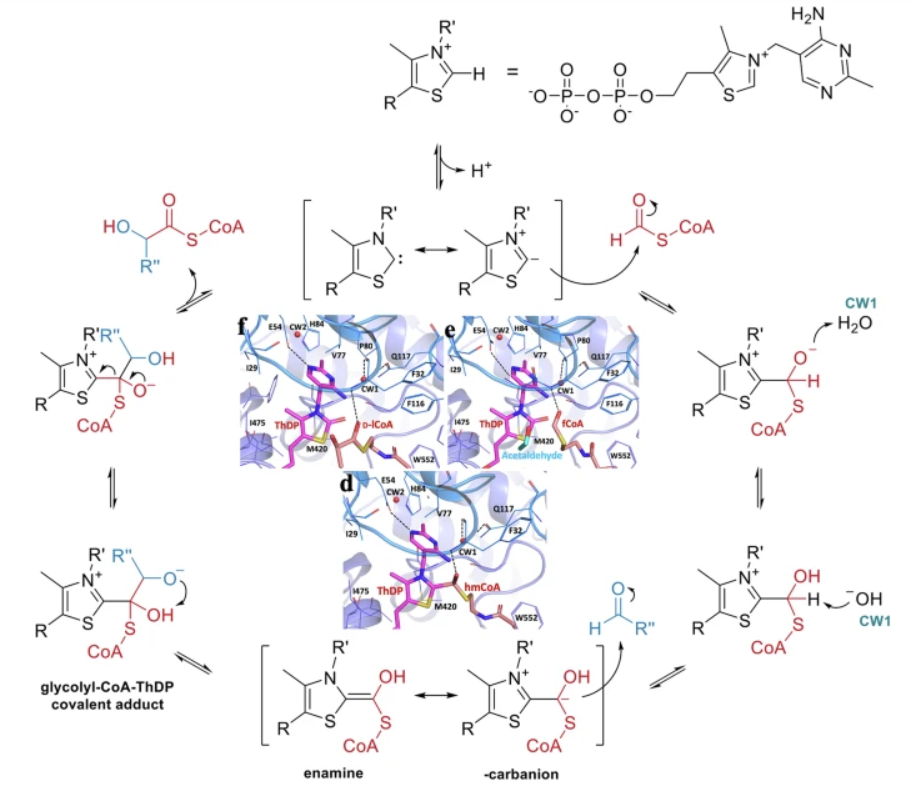
After purifying the expressed proteins, some members of the international team characterized the enzymes biochemically. Others produced crystals of five enzymes separately and in complexes with acyl-CoA substrates, ThDP cofactor, and ADP. They X-rayed the crystals, diffracted to 1.70–2.70 Å, at beamlines 19-ID – the Structural Biology Center (SBC) – and 23-ID-B – the National Institute of General Medical Sciences and National Cancer Institute Structural Biology Facility (GM/CA) – of the Advanced Photon Source (APS) at Argonne National Laboratory.
The crystal structures obtained from the X-ray data revealed what computer-predicted models could not: a flexible loop on the C terminus that locked on the cofactor and kept it bound to the enzyme’s active site. When the substrate was added, the loop closed the active site, stabilizing the cofactor and enabling the transfer of the formate compound to the substrate.
These reactions were observed in intermediate states, which also revealed unexpected products where the carbon atoms were added to the wrong position: “off” pathways that impeded normal process and reduced product yield.
The team’s findings can serve as the basis for much future research. Scientists can mutate the sequence of amino acids to produce stronger binders. They can experiment with different subfamilies of HACL/S enzymes that have longer or shorter C terminal regions. They can investigate whether ADP can stabilize particular states of the enzyme to accelerate its reaction. They can search for enzymes that avoid interfering off pathways. They can create enzymes that don’t obstruct the bacteria’s own metabolic pathways.
The team characterizes their findings as “just the beginning.” Eventually, they hope, their research can lead to new products such as stand-alone biofuels, enzymes that block the development of cancer cells, and bacteria that can gobble up plastic and spit out recyclable monomers.
And, of course, there’s the added benefit of using atmospheric carbon dioxide and methane precursors that can be converted to formate and be used by HACL/S enzymes to produce new products for the circular economy rather than storing it underground or converting it into rocks. One more cherry on top: reducing the carbon dioxide in the atmosphere releases oxygen needed to change methane into formate—a necessary ingredient for the biocatalysis to occur in the first place. – Judy Myers
_____________________________________________________________________________
See: Y. Kim1,2, S.H. Lee3, P. Gade2, M. Natterman4, N. Maltseva2, M. Endries1, J. Chen3, P. Wichmann3, Y. Hu3, D.G. Marchal4, Y. Yoshikuni5, T.J. Erb4,6, R. Gonzalez3, K. Michalska1,2, A. Joachimiak1,2, “Revealing reaction intermediates in one-carbon elongation by thiamine diphosphate/CoA-dependent enzyme family,” Commun Chem 7 160 (2024)
Author affiliations: 1Argonne National Laboratory; 2University of Chicago; 3University of South Florida, 4Max Planck Institute for Terrestrial Microbiology, 5Lawrence Berkeley National Laboratory, 6Center for Synthetic Microbiology
We truthfully thank the members of the SBC at Argonne National Laboratory, especially Alex Lavens for his help with setting beamline and data collection, Paula Bulaon and Michelle Radford for help in preparing this manuscript. Results in this report are derived from work performed through the eBERlight and Structural Biology Center programs, funded by the U.S. Department of Energy, Office of Biological and Environmental Research at the Advanced Photon Source, a U.S. DOE Office of Science User Facility, supported by the U.S. DOE, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. The work (proposal: https://doi.org/10.46936/10.25585/60001266) conducted by the U.S. Department of Energy Joint Genome Institute (https://ror.org/04xm1d337), a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy operated under Contract No. DE-AC02-05CH11231 and U.S. Department of Energy Contract No. DE-EE0008499.
The U.S. Department of Energy's APS at Argonne National Laboratory is one of the world’s most productive x-ray light source facilities. Each year, the APS provides high-brightness x-ray beams to a diverse community of more than 5,000 researchers in materials science, chemistry, condensed matter physics, the life and environmental sciences, and applied research. Researchers using the APS produce over 2,000 publications each year detailing impactful discoveries and solve more vital biological protein structures than users of any other x-ray light source research facility. APS x-rays are ideally suited for explorations of materials and biological structures; elemental distribution; chemical, magnetic, electronic states; and a wide range of technologically important engineering systems from batteries to fuel injector sprays, all of which are the foundations of our nation’s economic, technological, and physical well-being.
Argonne National Laboratory seeks solutions to pressing national problems in science and technology. The nation's first national laboratory, Argonne conducts leading-edge basic and applied scientific research in virtually every scientific discipline. Argonne researchers work closely with researchers from hundreds of companies, universities, and federal, state and municipal agencies to help them solve their specific problems, advance America's scientific leadership and prepare the nation for a better future. With employees from more than 60 nations, Argonne is managed by UChicago Argonne, LLC, for the U.S. DOE Office of Science.
The U.S. Department of Energy's Office of Science is the single largest supporter of basic research in the physical sciences in the United States and is working to address some of the most pressing challenges of our time. For more information, visit the Office of Science website.
