 Transfer RNAs (tRNAs) are RNA molecules used by all forms of life, from bacteria to plants to humans, to transfer amino acids to growing protein molecules that have been coded by DNA and transcribed into messenger RNA (mRNA) for translation by ribosomes into proteins. This is one of the most basic, crucial processes of life.
Transfer RNAs (tRNAs) are RNA molecules used by all forms of life, from bacteria to plants to humans, to transfer amino acids to growing protein molecules that have been coded by DNA and transcribed into messenger RNA (mRNA) for translation by ribosomes into proteins. This is one of the most basic, crucial processes of life.
However, tRNAs do more than just perform this essential function and are known to have regulatory roles in translation, transcription, stress response, and even immunity, via specific interactions with a wide array of cellular molecules. Disruption of these interactions has also been shown to be associated with some types of neurological disease and cancer, making it critical to understand how proteins in the cell recognize tRNAs.
Many different proteins have been shown to interact with tRNAs via known protein structural motifs. One of these is the oligonucleotide/oligosaccharide-binding (OB)-fold that has a highly conserved β-barrel structure found in organisms across all domains of life. However, the details of its interactions with tRNA are not completely understood. Recent research from a team at the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) has provided new, previously unrecognized, insights into how the OB-fold recognizes the 3’ tail of tRNA molecules and how these interactions impact the function of tRNAs.
The research team used X-ray diffraction data collected at the South East Regional Collaborative Access Team (SER-CAT) beamline 22-ID of the Advanced Photon Source, a U.S. Department of Energy (DOE) user facility at DOE’s Argonne National Laboratory.
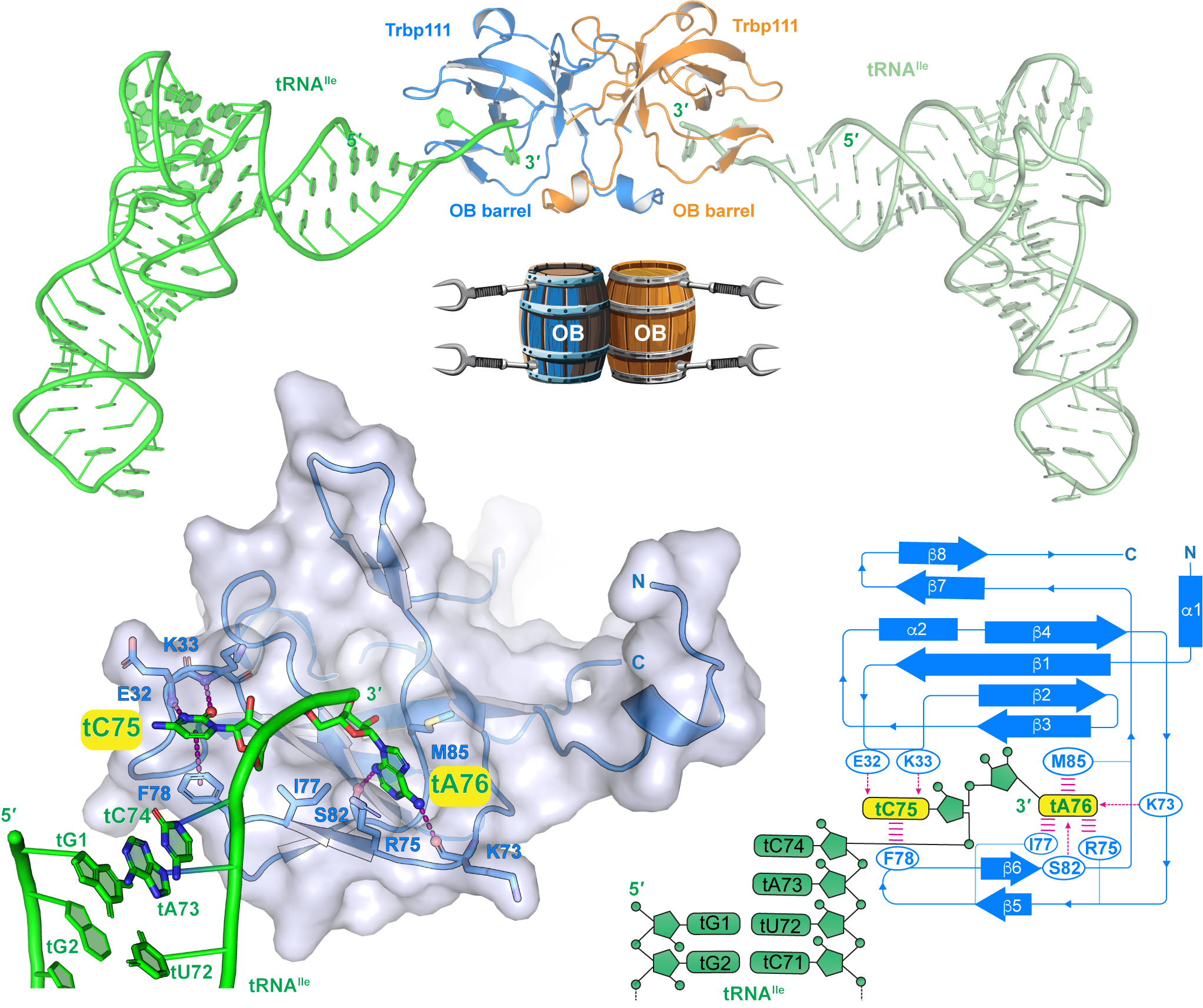
The protein structure work in this project focused on a superfamily of proteins called the tRNA binding domain (TRBD) family. These proteins interact with tRNAs via an OB-fold domain that consists of five β strands that form a barrel structure with an α-helical cap. TRBD proteins are found in many different organisms and, while they don’t always have high levels of amino acid sequence conservation, they all contain the OB-fold.
This work started with a TRBD protein from the bacteria Aquifex aeolicus called Trbp111 that is known to bind to many tRNAs. Solution of a new 2.3 Å crystal structure of Trbp111 showed that the protein forms an unusually stable homodimer with the two β-barrels stabilized by a very strong dimer interface. This is consistent with what is known about Trbp111, as A. aeolicus thrives at high temperatures (~90°C) and is also resistant to many common laboratory protein denaturing procedures, suggesting that this type of stable interface may provide a model for artificial protein design and structure-based drug design efforts.
The next step in the research was to analyze the interaction of Trbp111 with a fluorescently labeled tRNA via fluorescent polarization assays. These experiments confirmed the binding of Trbp111 to the tRNA and provided an initial demonstration that Trbp111 binds to the tRNA via its 3’ tail with a micromolar affinity that is consistent with what is known about its biological activity. The 2.6Å crystal structure of the Trbp111 - tRNA complex produced by co-crystallization showed that each subunit of the dimer binds to one tRNA via its 3’ tail.
Further competition assays using tRNAs with deletions of various parts of the molecule, such as the elbow and T-loop that had been implicated in previous studies, confirmed that the interactions are entirely mediated by the 3’ tail. Addition of more nucleotides or phosphate groups to the tail also abolished or diminished binding, showing that Trbp111 precisely recognizes the 3’ terminus of mature tRNAs.
Comparing the structure of A. aeolicus TRBD to that of a yeast TRBD protein called Arc1p with a similar OB-fold showed that Arc1p has the same mode of tRNA binding but is more permissive with regard to contacts at the 3’ tail and also interacts with the tRNA through another region in the middle of the protein.
These data provide valuable insights into how TRBD proteins interact with tRNAs to perform their essential functions. The authors hypothesize that the interactions could play a role in chaperoning the tRNA during folding to avoid entanglement or aggregation, protecting the 3’ end from exonuclease digestion or chemical degradation in harsh environments, and may facilitate trafficking of tRNAs to the ribosome or other cellular locations. These roles are consistent with many of the known functions of tRNAs and this new research provides important information for understanding basic cellular processes and for designing treatments. – Sandy Field
_____________________________________________________________________________
See: A.U. Juru1, R. Ghirlando1, J. Zhang1, “Structural basis of tRNA recognition by the widespread OB fold,” Nat Commun 15 6385 (2024)
Author affiliations: 1National Institute of Diabetes and Digestive and Kidney Diseases
We thank I. Skeparnias for providing human mascRNA, I. Botos for computational support, G. Piszczek and D. Wu for support in biophysical analyses, and C. Stathopoulos, C. Bou-Nader, K. Suddala, I. Skeparnias, A.-N. Shaukat, and A. Brasington for discussions. This work was supported by the Intramural Research Program of the NIH, the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) (ZIADK075136 to J.Z.), and an NIH Deputy Director for Intramural Research (DDIR) Challenge Award to J.Z. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science user facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
Data were collected at Southeast Regional Collaborative Access Team (SER-CAT) 22-ID-D (or 22-ID-E) beamline at the Advanced Photon Source, Argonne National Laboratory. SER-CAT is supported by its member institutions, equipment grants (S10_RR25528, S10_RR028976 and S10_OD027000) from the National Institutes of Health, and funding from the Georgia Research Alliance.
The U.S. Department of Energy's APS at Argonne National Laboratory is one of the world’s most productive x-ray light source facilities. Each year, the APS provides high-brightness x-ray beams to a diverse community of more than 5,000 researchers in materials science, chemistry, condensed matter physics, the life and environmental sciences, and applied research. Researchers using the APS produce over 2,000 publications each year detailing impactful discoveries and solve more vital biological protein structures than users of any other x-ray light source research facility. APS x-rays are ideally suited for explorations of materials and biological structures; elemental distribution; chemical, magnetic, electronic states; and a wide range of technologically important engineering systems from batteries to fuel injector sprays, all of which are the foundations of our nation’s economic, technological, and physical well-being.
Argonne National Laboratory seeks solutions to pressing national problems in science and technology. The nation's first national laboratory, Argonne conducts leading-edge basic and applied scientific research in virtually every scientific discipline. Argonne researchers work closely with researchers from hundreds of companies, universities, and federal, state and municipal agencies to help them solve their specific problems, advance America's scientific leadership and prepare the nation for a better future. With employees from more than 60 nations, Argonne is managed by UChicago Argonne, LLC, for the U.S. DOE Office of Science.
The U.S. Department of Energy's Office of Science is the single largest supporter of basic research in the physical sciences in the United States and is working to address some of the most pressing challenges of our time. For more information, visit the Office of Science website.
