Plastic waste is one of the world’s largest environmental challenges. One type of plastic, called Poly(ethylene terephthalate) or PET, makes up 12% of solid waste on Earth. A new study, which involved experiments performed at the U.S. Department of Energy’s Advanced Photon Source (APS), demonstrates a robust closed-loop recycling process for PET using an enzyme designed with artificial intelligence. If adopted, this technology could have positive impacts on the plastic recycling industry, reduce plastic waste, and help prevent further environmental devastation. The results of this research were published in the journal Nature.
Plastic is a tremendously versatile material that touches just about every element of our lives. It also creates a lot of waste. Over 322 million metric tons of plastic are produced annually, and the average person now disposes of around 115 pounds of plastic a year. This plastic waste is a major environmental burden that contaminates our water system and harms the health of humans and other species.
One way to reduce plastic waste is to improve our ability to break down plastics and recycle them. A new study demonstrates how an enzyme engineered with the help of machine learning can be used to efficiently recycle a common type of plastic called poly(ethylene terephthalate) or PET. Previously discovered PET-degrading enzymes only work well in a narrow range of temperatures and pHs and lose activity over time, which makes them not very useful for PET recycling. The researchers in this study sought to engineer more stable and effective enzymes using a structure-based machine learning algorithm.
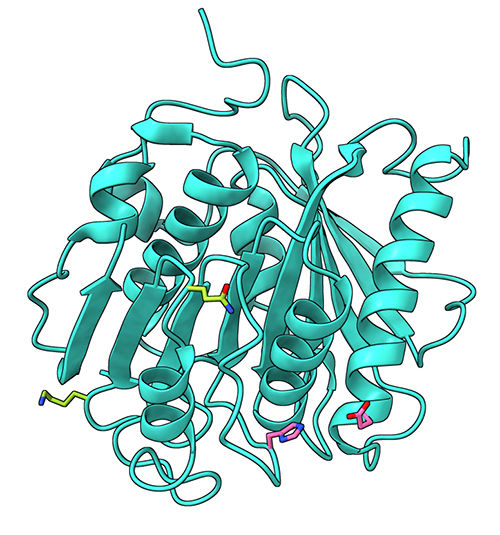
The research team started with the structure of the PET-degrading enzyme (PETase) from the bacterium Ideonella sakaiensis as well as the structures of two previously engineered variants of this enzyme (ThermoPETase and DuraPETase). Using a machine learning algorithm that had been trained with over 19,000 protein structures, the team identified 159 possible PETase variants with mutations that could improve enzyme stability and activity. With further analysis, the researchers were able to identify the most improved variant, which they named FAST-PETase (functional, active, stable, and tolerant PETase) (Fig. 1).
Experiments found that FAST-PETase was much more efficient at breaking down a thin film of PET compared to ThermoPETase. To obtain an insight into how mutations suggested by the machine learning algorithm improve the enzyme activity, the team solved the crystal structure at 1.4 Å using x-ray diffraction data collected at the National Institute of General Medical Sciences and National Cancer Institute (GM/CA-XSD) 23-ID-B beamline in the APS, an Office of Science user facility at Argonne National Laboratory. A three-dimensional, atomic-resolution structure enabled the researchers to experimentally confirm how the mutations make FAST-PETase stable and efficient compared to the wild-type enzyme.
The researchers then tested FAST-PETase’s ability to break down 51 hole-punched samples of PET used in packaging for food, beverages, medications, office supplies, household goods, and cosmetics. FAST-PETase fully degraded all of these samples within a week and in as little as 24 hours. The process was also easily scalable—treating larger pieces of plastic just required using more enzyme.
Some PET materials, like plastic water bottles, have parts that are highly crystalline in structure, making them less susceptible to being degraded by enzymes, including FAST-PETase. However, the team found that when a bottle was melted first, FAST-PETase could completely degrade it in less than two weeks at 50 °C.
The researchers also determined that FAST-PETase could rapidly break down polyester fabric, another form of PET, suggesting that it could be used for recycling clothes and other textiles.
Importantly, the research showed that monomers collected from breaking down PET with FAST-PETase could be repolymerized into new materials with a very high yield and purity. The entire cycle of degradation and repolymerization could be completed in just a few days. An added advantage is that the repolymerized plastic is clear, which means mixed-colored plastic can be recycled without sorting.
Together, this study demonstrates a new method for closed-loop PET enzymatic recycling at an industrial scale. If adopted, this technology could make a major dent in reducing the world’s plastic waste, which would have a whole host of downstream ecological benefits. ― Summer Allen
See: Hongyuan Lu1, Daniel J. Diaz1, Natalie J. Czarnecki1, Congzhi Zhu1, Wantae Kim1, Raghav Shroff1,2, Daniel J. Acosta1, Bradley R. Alexander1, Hannah O. Cole1, Yan Zhang1, Nathaniel A. Lynd1, Andrew D. Ellington1, and Hal S. Alper1*, “Machine learning-aided engineering of hydrolases for PET depolymerization,” Nature 604, 662 (28 April 2022). DOI: 10.1038/s41586-022-04599-z
Author affiliations: 1The University of Texas at Austin, 2DEVCOM ARL-South
Correspondence: * halper@che.utexas.edu
This work was financed under research agreement no. EM10480.26/ UTA16-000509 between the ExxonMobil Research and Engineering Company and The University of Texas at Austin. Sequencing was conducted at the Genomic Sequencing and Analysis Facility (RRID no. SCR_021713), SEM was conducted at the Microscopy and Imaging Facility (RRID no. SCR_021756) at the UT Austin Center for Biomedical Research Support, and AFM analysis was conducted at the Texas Materials Institute at UT Austin. N.A.L. and C.Z. thank the Welch Foundation for partial support of this research (Grant #F-1904). The crystallography study is supported by a grant from the National Institutes of Health (no. GM104896 to Y.J.Z.). GM/C-XSD has been funded by the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006, P30GM138396). We acknowledge the Texas Advanced Computing Center at The University of Texas at Austin for providing deep learning resources for neural network predictions and analysis that have contributed to the research results reported in this paper. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science user facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
The U.S. Department of Energy's APS at Argonne National Laboratory is one of the world’s most productive x-ray light source facilities. Each year, the APS provides high-brightness x-ray beams to a diverse community of more than 5,000 researchers in materials science, chemistry, condensed matter physics, the life and environmental sciences, and applied research. Researchers using the APS produce over 2,000 publications each year detailing impactful discoveries and solve more vital biological protein structures than users of any other x-ray light source research facility. APS x-rays are ideally suited for explorations of materials and biological structures; elemental distribution; chemical, magnetic, electronic states; and a wide range of technologically important engineering systems from batteries to fuel injector sprays, all of which are the foundations of our nation’s economic, technological, and physical well-being.
Argonne National Laboratory seeks solutions to pressing national problems in science and technology. The nation's first national laboratory, Argonne conducts leading-edge basic and applied scientific research in virtually every scientific discipline. Argonne researchers work closely with researchers from hundreds of companies, universities, and federal, state and municipal agencies to help them solve their specific problems, advance America's scientific leadership and prepare the nation for a better future. With employees from more than 60 nations, Argonne is managed by UChicago Argonne, LLC, for the U.S. DOE Office of Science.
The U.S. Department of Energy's Office of Science is the single largest supporter of basic research in the physical sciences in the United States and is working to address some of the most pressing challenges of our time. For more information, visit the Office of Science website.
