The ability to distinguish between what’s safe and what’s unsafe protects us from disastrous consequences. Now, researchers from the Yale University School of Medicine and Yale University have discovered that this is true even on a cellular level. They found that in a certain type of brain cancer, mutations in a cellular receptor impair its ability to discriminate between a molecular signal that promotes normal cell differentiation and another biomolecule that uses the same receptor to promote cancer—thereby interfering with the former molecule’s ability to do its health-enhancing job. The scientists’ findings, using crystallographic data obtained from research at the U.S. Department of Energy’s Advanced Photon Source (APS), have profound implications for our understanding of a basic mechanism of cancer origination, as well as how to treat specific conditions, such as glioblastoma and lung cancer.
Glioblastoma, an aggressive brain tumor, occurs especially in the frontal and temporal lobes, areas that control higher-level executive functions and language recognition. To date, targeted therapies that have been effective in other cancers have not proven effective in glioblastoma, making it a subject of focused study.
The current work zeroed in on a specific receptor on the surface of human cells. Receptors are like antennae that detect signaling biomolecules in the extracellular environment. These signaling molecules tell the cells what to do. In glioblastoma, the epidermal growth factor receptor (EGFR) can detect several different signaling biomolecules, including epiregulin (EREG) and epidermal growth factor (EGF). When either of these biomolecules binds to the receptor, the receptor changes shape, with two molecules coming together in a structure called a dimer.
When EGF binds to the receptor, the resulting dimer is strong and symmetrical. However, as this team learned from research in 2017, when EREG binds, the receptor dimer is weaker and asymmetrical. More importantly, they discovered that this difference in the strength of the dimer specifies the type of signal that is sent to the cell. A weak dimer signals cell differentiation, whereas a strong dimer signals cell proliferation, which can promote cancer.
The current work by this group expands on that finding in an important way, by showing that mutations in the EGFR found in glioblastoma cause the receptor to make aberrantly strong dimers when EREG binds, rather than the weak dimers that this signaling biomolecule normally produces in the non-mutated receptor. By creating the wrong dimer, the mutations cause the receptor to send the wrong message to the cell, telling it to proliferate rather than differentiate in response to EREG.
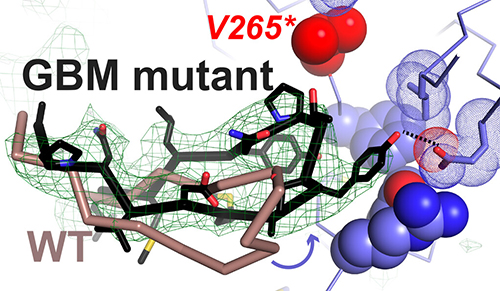
Using the National Institute of General Medical Sciences and National Cancer Institute (GM/CA-XSD) structural biology facility x-ray beamline 23-ID-B at the APS, the scientists produced two crystal structures that revealed at the atomic level how the glioblastoma mutations transform a weak EREG-bound dimer into a strong dimer (Fig. 1). The first crystal structure revealed that an R84K mutation breaks up a small cluster of protein sidechains that normally keep the EREG-induced dimer asymmetrical, thereby forcing it into a strong and symmetric dimer structure. The second crystal structure revealed that the most common A265V mutation in glioblastoma instead allows key sidechains to redock to form a strong dimer.
These findings reveal an important characteristic of EGFR mutations in glioblastoma that may explain why inhibitors of this receptor have not been successful. In other cancers, such as lung cancer, EGFR mutations simply turn the receptor “on,” causing it to drive cell proliferation. Treatment consists of administering an inhibitor that turns this switch off. In glioblastoma, by contrast, the EGFR mutations change how the receptor signals—possibly before tumor development—rather than driving tumor development directly. The current research suggests that inhibitors may have failed in glioblastoma because the mutated EGFR promotes tumor development, but not necessarily progression.
However, the team’s structural models showing how EGFR mutations influence cell signaling specificity may suggest new ways to “correct” EGFR signaling when it has gone awry: creating a biologic that could conformationally “cure” the receptor and make it good again. For instance, an antibody could be developed to reintroduce asymmetry into EREG-induced dimers of mutated EGFR and weaken them to promote cell differentiation. It is not clear whether this would be helpful in glioblastoma once the tumor has established itself but could be important in other cancers and other contexts.
The team is already looking into whether this mechanism of altered EGFR signaling specificity is specific for glioblastoma, or whether extracellular EGFR mutations do the same or different things in other cancers. There are also additional mutations in glioblastoma that the team hasn’t yet studied. They want to know whether those do the same thing as the mutations they studied here. ― Judy Myers
See: Chun Hu1,2, Carlos A. Leche II1,2, Anatoly Kiyatkin1,2, Zhaolong Yu1, Steven E. Stayrook1,2, Kathryn M. Ferguson1,2, and Mark A. Lemmon1,2*, “Glioblastoma mutations alter EGFR dimer structure to prevent ligand bias,” Nature 602, 518 (17 February 2022). DOI: 10.1038/s41586-021-04393-3
Author affiliations: 1Yale University School of Medicine, 2Yale University
Correspondence: * mark.lemmon@yale.edu
This work was supported by National Cancer Institute (NCI) grant R01-CA198164 (to M.A.L. and K.M.F.). GM/CA-XSD is funded by the NCI (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006, P30GM138396). The Eiger 16M detector at GM/CA-XSD was funded by National Institutes of Health grant S10 OD012289. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science user facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
The U.S. Department of Energy's APS at Argonne National Laboratory is one of the world’s most productive x-ray light source facilities. Each year, the APS provides high-brightness x-ray beams to a diverse community of more than 5,000 researchers in materials science, chemistry, condensed matter physics, the life and environmental sciences, and applied research. Researchers using the APS produce over 2,000 publications each year detailing impactful discoveries, and solve more vital biological protein structures than users of any other x-ray light source research facility. APS x-rays are ideally suited for explorations of materials and biological structures; elemental distribution; chemical, magnetic, electronic states; and a wide range of technologically important engineering systems from batteries to fuel injector sprays, all of which are the foundations of our nation’s economic, technological, and physical well-being.
Argonne National Laboratory seeks solutions to pressing national problems in science and technology. The nation's first national laboratory, Argonne conducts leading-edge basic and applied scientific research in virtually every scientific discipline. Argonne researchers work closely with researchers from hundreds of companies, universities, and federal, state and municipal agencies to help them solve their specific problems, advance America's scientific leadership and prepare the nation for a better future. With employees from more than 60 nations, Argonne is managed by UChicago Argonne, LLC, for the U.S. DOE Office of Science.
The U.S. Department of Energy's Office of Science is the single largest supporter of basic research in the physical sciences in the United States and is working to address some of the most pressing challenges of our time. For more information, visit the Office of Science website.
