It’s getting to be more than a little old: Every time it seems that SARS-CoV-2, the virus that causes COVID-19, begins to wane, giving the entire globe a glimmer of hope that this pandemic will end once and for all, a new mutant starts a new wave. It turns out, SARS-CoV-2 has a mutagenic superpower, boasting an RNA-dependent RNA polymerase (RdRp) without the proofreading domain typical of DNA polymerases and resulting in the high mutation rate believed to facilitate the virus’ ability to rapidly adapt. And SARS-CoV’s RdRp appears to be the fastest of the viral RdRps and has an error rate an order of magnitude higher than other viral RdRps. That spells trouble for the virus’ ability to replicate, because while some mutations confer adaptability, too many can reduce viability. SAR-Co-2’s answer to this conundrum is a proofreading exonuclease, which fixes the viral genome enough to keep the pathogen happily replicating. In this study, the researchers took aim at the exonuclease using high-brightness x-rays from the U.S. Department of Energy’s Advanced Photon Source (APS) to gather data that allowed them to determine the structure of the exonuclease. The findings, published in the Proceedings of the National Academy of Sciences of the United States of America, may provide the basis for the future development of therapeutics that target the exonuclease, attenuating SARS-CoV-2’s proofreading capacity and thereby potentially improving patient outcomes.
Beyond the global pandemic, the coronaviruses are noteworthy because they have the largest known RNA virus genomes, in the ballpark of 27 to 32 kilobases, which is up to twice the length of the next largest. That means there is a lot that can potentially go wrong during replication, necessitating a strong proofreader. Researchers established the importance of the exonuclease through a series of studies. For example, coronaviruses with an inactivated exonuclease was less virulent. In another study, viruses with inactivate exonuclease were more sensitive to nucleoside analogs that target RdRps, suggesting that the function of the exonucleases is to remove problematic nucleotides that inhibit viral replication.
Another set of experiments involved sofosbuvir (sold under the brand name Sovaldi), a drug that has been shown to have antiviral activity against SARS-CoV-2 in the laboratory. Sofosbuvir is a chain terminator, which is an agent that stops the extension of a nucleic acid chain in its tracks. The researchers mixed together all the ingredients needed for RNA chain extension, adding in sofosbuvir, and observed the expected chain termination. However, adding in the active exonuclease rescued the chain extension, allowing RdRp to more faithfully replicate the viral genome.
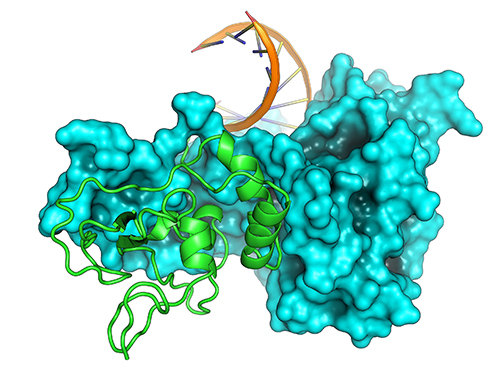
To get a better idea of how the exonuclease works, the researchers used x-ray crystallography at the Northeastern Collaborative Access Team (NE-CAT) x-ray beamline 24-ID-C to solve a 1.6-angstrom structure of the exonuclease in complex with nsp10. Previous research showed that the SARS-CoV-2 exonuclease proofreader forms a complex with nonstructural protein 10 (nsp10) to gain activity. The structure reveals a curved surface with basic residues around the active site, with a hand over fist type structure, with the exonuclease as the hand and nsp10 as the fist. The positively charged lysine residues provide a perfect chemistry to bind the template strand of RNA, which is strongly negatively charged. The researchers used modeling software to get a sense of how the exonuclease-nsp10 structure engages RNA using data from a previously determined structure of a Lassa virus exonuclease domain bound to double stranded RNA. The modeling supported the hypothesis that the basic patch on the exonuclease positions the RNA substrate for the reaction that cleaves out residues that have been mis-incorporated.
To assess the impact of protein movement on exonuclease behavior, the researchers performed molecular dynamics simulations. The simulation showed remarkable flexibility of the exonuclease when on its own, suggesting that the nsp10 contributes the stability that facilitates RNA binding and exonuclease activity.
Together, this research provides insights into how viruses can be both highly mutagenic and highly pathogenic, but also offers pathways for the development of drugs that may be able to curtail these viral mechanisms. And the sooner, the better. ― Erika Gebel Berg
See: Nicholas H. Moeller1, Ke Shi1, Özlem Demir2, Christopher Belica1, Surajit Banerjee3, Lulu Yin1, Cameron Durfee1, Rommie E. Amaro2 , and Hideki Aihara1*, “Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN,” Proc. Natl. Acad. Sci. USA 119(9), e2106379119 (2022). DOI: 10.1073/pnas.2106379119
Author affiliations: 1University of Minnesota, 2University of California, San Diego, 3Cornell University
Correspondence: * aihar001@umn.edu
This work was supported by grants from the US NIH [National Institute of General Medical Sciences (NIGMS) R35-GM118047 to H.A., R01-GM132826 to R.E.A., and National Cancer Institute P01-CA234228 to R.E.A. and H.A.], NSF Rapid Response Research MCB-2032054, an award from the RCSA Research Corp., and a University of California San Diego Moores Cancer Center 2020 SARS-COV-2 seed grant to R.E.A. This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the US NIH (NIGMS P30 GM124165). The Pilatus 6M detector on 24-ID-C beamline is funded by NIH Office of Research Infrastructure Programs High-End Instrumentation Grant S10 RR029205. This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357, and those of the Minnesota Supercomputing Institute. We are grateful for the efforts of the Texas Advanced Computing Center Frontera team and for the computing time made available through a Director’s Discretionary Allocation (made possible by NSF Award OAC-1818253).
This research used resources of the Advanced Photon Source, a U.S. DOE Office of Science user facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
The U.S. Department of Energy's APS at Argonne National Laboratory is one of the world’s most productive x-ray light source facilities. Each year, the APS provides high-brightness x-ray beams to a diverse community of more than 5,000 researchers in materials science, chemistry, condensed matter physics, the life and environmental sciences, and applied research. Researchers using the APS produce over 2,000 publications each year detailing impactful discoveries, and solve more vital biological protein structures than users of any other x-ray light source research facility. APS x-rays are ideally suited for explorations of materials and biological structures; elemental distribution; chemical, magnetic, electronic states; and a wide range of technologically important engineering systems from batteries to fuel injector sprays, all of which are the foundations of our nation’s economic, technological, and physical well-being.
Argonne National Laboratory seeks solutions to pressing national problems in science and technology. The nation's first national laboratory, Argonne conducts leading-edge basic and applied scientific research in virtually every scientific discipline. Argonne researchers work closely with researchers from hundreds of companies, universities, and federal, state and municipal agencies to help them solve their specific problems, advance America's scientific leadership and prepare the nation for a better future. With employees from more than 60 nations, Argonne is managed by UChicago Argonne, LLC, for the U.S. DOE Office of Science.
The U.S. Department of Energy's Office of Science is the single largest supporter of basic research in the physical sciences in the United States and is working to address some of the most pressing challenges of our time. For more information, visit the Office of Science website.
