Currently, more than 460 million adults worldwide live with diabetes and its complications. That number is expected to grow to 700 million adults by 2045. Unfortunately, available treatments for type 2 diabetes, which make up approximately 90 percent of adult cases, can have serious adverse effects including gastrointestinal problems, hypoglycemia, increased cholesterol, and even heart failure. A new study carried out by a multinational, multi-institution research group, which involved collecting x-ray diffraction data at the U.S. Department of Energy’s Advanced Photon Source (APS), provides important insights into the structure of the human leukotriene B4 receptor 1, which is thought to play an important role in type 2 diabetes. This research, published in Nature Communications, could result in new treatments for diabetes as well as other inflammatory conditions like asthma, inflammatory bowel disease, rheumatoid arthritis, and cancer.
Inflammation is a key hallmark of many diseases, including diabetes. Several studies implicate a protein called the leukotriene B4 receptor 1 (BLT1), which is found in white blood cells, as being involved in inflammatory processes that cause disease. Research also suggests that blocking BLT1 activity may be a potent strategy for combating inflammatory diseases like type 2 diabetes. For example, one study found that removing BLT1 using genetic engineering prevented obese mice from developing insulin resistance, a precursor to diabetes.
Unfortunately, attempts to develop drugs that target BLT1 have been unsuccessful thus far with potential candidates showing poor efficacy and adverse side effects. One reason why these attempts have not been successful may be that the compounds developed may not have been able to selectively bind BLT1 and block its activity.
In order to learn more about BLT1 and how it might be better targeted by candidate drugs, a multi-institution group conducted a series of experiments designed to reveal the structure of human BLT1 and to determine how various molecules interact with the receptor.
First, the researchers used macromolecular x-ray crystallography to create a detailed, three-dimensional structure of human BLT1 while it was bound to MK-D-046, a selective BLT1 blocker that was developed by Merck & Co. as a candidate for type 2 diabetes treatment. These data were collected at the National Institute of General Medical Sciences and National Cancer Institute (GMCA-XSD) 23-ID-B x-ray beamline at the APS (the APS is a DOE Office of Science user facility at Argonne National Laboratory).
The results showed that human BLT1 has a structure similar to many other receptors that span cell membranes and includes an internal channel pocket. The crystal structure also allowed the researchers to determine exactly how MK-D-046 “docks” within the receptor’s pocket.
Using specialized software, the researchers modeled how leukotriene B4 (LTB4) likely docks with the human BLT1. LTB4 is a naturally occurring binding partner for the receptor and is thought to play a protective role in viral infections and in some forms of lung injury.
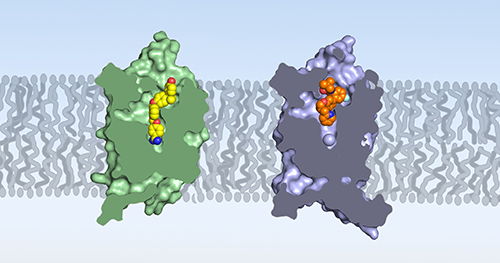
The researchers also compared the structure of human BLT1 to the previously reported structure of guinea pig BLT1, which had been the previous best model for understanding drug interaction with the human receptor (Fig. 1). They found significant differences in the receptor structures, including at sites where potential drugs would bind. This is despite the fact that the amino acid sequences of human BLT1 and guinea pig BLT1 are nearly 74% similar.
To test which differences between the human and guinea pig BLT1 are likely to be most important for drug binding, the researchers mutagenized particular amino acids within human BLT1 so they matched sites from guinea pig BLT1. They then tested how well MK-D-046 blocked the mutated human BLT1. They found that certain amino acid substitutions significantly decreased the ability of MK-D-046 to inhibit human BLT1 activity, thus gaining an understanding of which amino acids are critical for high-affinity binding of drugs to human BLT1.
Together, these results suggest that the structure of human BLT1 will be useful for designing compounds that target BLT1. By presenting a detailed structure of human BLT1, the results from this study may one day lead to the development of more effective methods for preventing and treating inflammatory diseases, including diabetes. ― Summer Allen
See: Nairie Michaelian1, Anastasiia Sadybekov1, Élie Besserer-Offroy2,3, Gye Won Han1, Harini Krishnamurthy4, Beata A. Zamlynny4, Xavier Fradera4, Phieng Siliphaivanh4, Jeremy Presland4, Kerrie B. Spencer4, Stephen M. Soisson4, Petr Popov5,6, Philippe Sarret2, Vsevolod Katritch1, and Vadim Cherezov1,6*, “Structural insights on ligand recognition at the human leukotriene B4 receptor 1,” Nat. Commun. 12, 2971 (2021). DOI: 10.1038/s41467-021-23149-1
Author affiliations: 1University of Southern California, 2Université de Sherbrooke, 3University of California at Los Angeles, 4Merck & Co., 5Skolkovo Institute of Science and Technology, 6Moscow Institute of Physics and Technology
Correspondence: * cherezov@usc.edu
This research was supported in parts by the National Institute of General Medical Sciences grant R35 GM127086, the GPCR Consortium, and the Canadian Institutes of Health Research (CIHR) grant FDN-148413. É.B.-O. is the recipient of research fellowships from the CIHR (MFE-164740) and the Fonds de recherche du Québec–Santé (255989) and a member of the California NanoSystem Institute. P.S. is the recipient of a Tier 1 Canada Research Chair in Neurophysiopharmacology of Chronic Pain and a member of the FRQ-S-funded Centre de recherche du CHUS. GM/CA-XSD has been funded in whole or in part with federal funds from the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006, P30GM138396). The Eiger-16M detector at GM/CA-XSD was funded by National Institutes of Health grant S10 OD012289. This research used resources of the Advanced Photon Source, a U.S. DOE Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357.
The U.S. Department of Energy's APS is one of the world’s most productive x-ray light source facilities. Each year, the APS provides high-brightness x-ray beams to a diverse community of more than 5,000 researchers in materials science, chemistry, condensed matter physics, the life and environmental sciences, and applied research. Researchers using the APS produce over 2,000 publications each year detailing impactful discoveries, and solve more vital biological protein structures than users of any other x-ray light source research facility. APS x-rays are ideally suited for explorations of materials and biological structures; elemental distribution; chemical, magnetic, electronic states; and a wide range of technologically important engineering systems from batteries to fuel injector sprays, all of which are the foundations of our nation’s economic, technological, and physical well-being.
Argonne National Laboratory seeks solutions to pressing national problems in science and technology. The nation's first national laboratory, Argonne conducts leading-edge basic and applied scientific research in virtually every scientific discipline. Argonne researchers work closely with researchers from hundreds of companies, universities, and federal, state and municipal agencies to help them solve their specific problems, advance America's scientific leadership and prepare the nation for a better future. With employees from more than 60 nations, Argonne is managed by UChicago Argonne, LLC, for the U.S. DOE Office of Science.
The U.S. Department of Energy's Office of Science is the single largest supporter of basic research in the physical sciences in the United States and is working to address some of the most pressing challenges of our time. For more information, visit the Office of Science website.
