Usually, eukaryotic cells organize the functions of life by compartmentalization inside lipid membranes, but some fast processes requiring great precision happen in compartments formed through liquid-liquid phase separation (LLPS), similar to the way oil droplets will separate in a water solution. LLPS has become increasingly recognized as an important biological process not only in normal physiology but in certain pathologies, including neurodegenerative diseases such as Alzheimer’s, amyotrophic lateral sclerosis, and Parkinson's. A more detailed understanding of LLPS could lead to better therapeutic approaches, but the blazingly fast microsecond-to-millisecond time scales on which LLPS happens as a solution changes from the one-phase to the two-phase regime have made characterization of the process extremely challenging. A team of researchers from a diverse range of institutions approached that challenge by using the high-brightness x-rays from the U.S. Department of Energy’s Advanced Photon Source (APS) to capture for the first time the kinetics of how LLPS happens in a biomolecular solution. The work appeared in Nature Communications.
The investigators from St. Jude Children's Research Hospital, the Max Planck Institute, the Illinois Institute of Technology, Washington University in St. Louis, and the National Institutes of Health focused on a prion-like domain called A1-LCD, the low-complexity domain of the hnRNPA1 protein, which has been implicated in neurodegenerative diseases in its mutated form. LLPS occurs when local conditions transform a single-phase solution to two phases. In the vast majority of biological systems, LLPS is initiated by a phenomenon known as nucleation. Previously, nucleation was thought to be largely dependent on perturbation of the equilibrium between the solvent and the biomolecules it contained, but the extreme time resolution achieved in this work reveals that the process is more complex than previously believed.
One factor used to facilitate A1-LCD phase separation in these experiments is the salt concentration of the solution, a parameter which can be used to control the “quench depth,” defined as the degree to which the solution is perturbed into the two-phase regime. The experimenters examined this process using fluorescence correlation spectroscopy to characterize A1-LCD molecules in six samples of different salt concentrations in equilibrium. As the salt concentration of the solution is increased, interactions among protein molecules increase and they begin to form into clusters.
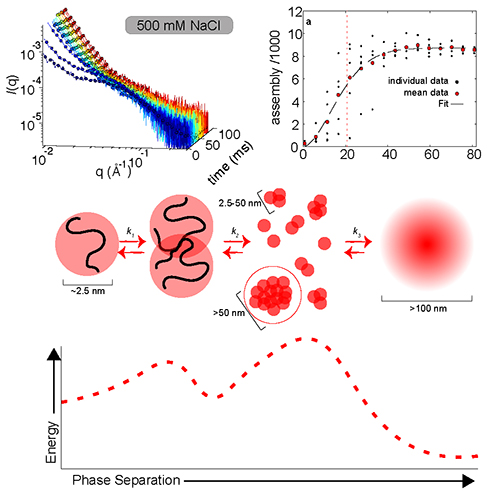
In studying the time-dependent process of LLPS, the research team used the technique of rapid-mixing, time-resolved small-angle x-ray scattering (TR-SAXS) at the Bio-CAT 18-ID-D beamline at the APS. The TR-SAXS techniques offer a detailed look into the earliest stages of the transition (Fig. 1). By combining data from two different microfluidic mixers, the investigators were able to conduct SAXS observations on a time scale from 80 microseconds to 80 milliseconds for the first time in the same experimental session. This provided key insights into the outstanding question of why the nucleation of some biomolecules can occur in fractions of a second, while others require hours to complete phase separation.
On the mesoscale, the A1-LCD phase separation seems to follow a classical homogeneous nucleation pattern. In this regime, the nucleation and assembly rate are determined by the quench depth, both slowing as quench depth decreases. Taking advantage of the power of SAXS to observe assembly also on the nanoscale, the researchers noticed that early assembly steps were unfavorable before additional monomers were added to form larger clusters. These findings indicate the presence of two distinct kinetic steps in the LLPS of A1-LCD, which introduce a time lag, noticeable for A1-LCD only with these high-precision measurements. This time lag likely contributes to the long time periods needed for phase separation in some proteins.
The work demonstrates that phase separation in some biological systems can be more complex than in simpler physical systems, which only require crossing the phase boundary. Since additional steps can occur at different timescales and conditions, equilibrium analysis is not enough to provide a full understanding of the phase separation process, and a careful kinetic analysis is necessary. Additional steps offer opportunities for cells to regulate phase separation, revealing the newly recognized biological usefulness of this process.
The small, bright beams of the APS Upgrade will make these kinds of SAXS experiments with microfluidic mixers much easier to do, with expected gains in time, resolution, and data quality. ― Mark Wolverton
See: Erik W. Martin1*, Tyler S. Harmon2, Jesse B. Hopkins3, Srinivas Chakravarthy3, J. Jeremías Incicco4, Peter Schuck5, Andrea Soranno4, and Tanja Mittag1**, “A multi-step nucleation process determines the kinetics of prion-like domain phase separation,” Nat. Commun. 12, 4513 (2021). DOI: 10.1038/s41467-021-24727-z
Author affiliations: 1St. Jude Children’s Research Hospital, 2The Max Planck Institute for the Physics of Complex Systems, 3Illinois Institute of Technology, 4Washington University in St. Louis, 5National Institutes of Health
Correspondence: * emartin@dewpointx.com, ** tanja.mittag@stjude.org
T.M. acknowledges funding by NIH grant R01NS121114, the St. Jude Children’s Research Hospital Research Collaborative on Membrane-less Organelles in Health and Disease, and by the American Lebanese Syrian Associated Charities. This work was supported by the Intramural Research Programs of the National Institute of Biomedical Imaging and Bioengineering, NIH, and resources supported by grant 9 P41 GM103622 from the National Institute of General Medical Sciences of the National Institutes of Health. Use of the Pilatus 3 1M detector was provided by grant 1S10OD018090-01 from NIGMS. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DEAC02- 06CH11357
The U.S. Department of Energy's APS is one of the world’s most productive x-ray light source facilities. Each year, the APS provides high-brightness x-ray beams to a diverse community of more than 5,000 researchers in materials science, chemistry, condensed matter physics, the life and environmental sciences, and applied research. Researchers using the APS produce over 2,000 publications each year detailing impactful discoveries, and solve more vital biological protein structures than users of any other x-ray light source research facility. APS x-rays are ideally suited for explorations of materials and biological structures; elemental distribution; chemical, magnetic, electronic states; and a wide range of technologically important engineering systems from batteries to fuel injector sprays, all of which are the foundations of our nation’s economic, technological, and physical well-being.
Argonne National Laboratory seeks solutions to pressing national problems in science and technology. The nation's first national laboratory, Argonne conducts leading-edge basic and applied scientific research in virtually every scientific discipline. Argonne researchers work closely with researchers from hundreds of companies, universities, and federal, state and municipal agencies to help them solve their specific problems, advance America's scientific leadership and prepare the nation for a better future. With employees from more than 60 nations, Argonne is managed by UChicago Argonne, LLC, for the U.S. DOE Office of Science.
The U.S. Department of Energy's Office of Science is the single largest supporter of basic research in the physical sciences in the United States and is working to address some of the most pressing challenges of our time. For more information, visit the Office of Science website.
