The economic, societal, and personal reverberations from the historic COVID-19 virus will be felt for years to come. As we continue to brace for new variants of SARS-CoV-2 and think about the potential for future pandemics, it is important that we develop a thorough understanding of the structures and reactivities of these deadly viruses so that we can be better prepared for the next global outbreak. To do this, we not only need to know the similarities between different viruses, but also their differences so that we can understand why some viruses only spread to a few thousand people whereas some have the ability to shut down the entire world. A team of researchers conducting experiments at the U.S. Department of Energy’s Advanced Photon Source (APS) are making significant strides in understanding the structures, active sites, and reactivity of SARS-CoV-2. At the early stage of the pandemic, the team characterized the reactivity and structure of a new subunit of SARS-CoV-2, a nidoviral RNA endoribonuclease known as NendoU. The insights gained through this research, published in the journal Protein Science, may lead to a better understanding of why SARS-CoV-2 is much more transmittable in comparison to other coronaviruses like SARS-CoV and MERS-CoV.
Since the start of the pandemic, we have learned much about the structure of SARS-CoV-2. In general, SARS-CoV-2 is a non-segmented positive-sense RNA virus that contains large replicase non-structural proteins (NSPs), followed by structural and accessory genes. Cleavage of the virus results in 15 or 16 NSPs that assemble into a large membrane-bound complex that exhibits enzymatic properties that are important for viral replication. These NSP complexes are segmented into 3 main domains: N-terminal domain, middle domain, and the C-terminal catalytic NendoU (or nidoviral RNA uridylate-specific endoribonuclease) domain. In general, the structures of NSPs are rare with a total of 8 NSP15 structures having been published prior to this study. NSP15 is particularly interesting because SARS-CoV-2 shares similar identity and similarity with SARS-CoV (88% and 95%, respectively) whereas MERS-CoV is significantly different with a 50% sequence identity and 65% similarity.
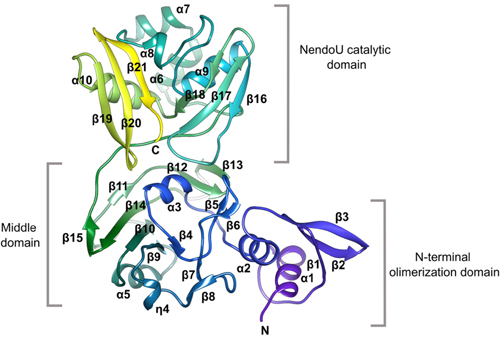
In this work, the research team reports new high-resolution structures of the NendoU domain of SARS-CoV-2 (Fig. 1) obtained at Structural Biology Center (SBC-XSD) beamline 19-ID at the DOE Office of Science’s APS at Argonne National Laboratory. In general, the N-terminal domain consists of an antiparallel ꞵ-sheet wrapped around two ɑ-helices while the middle domain is formed by 10 ꞵ-strands organized in three hairpins, a mixed ꞵ-sheet, and three short helices. Of particular interest is the structure of the NendoU catalytic domain which is composed of two antiparallel ꞵ-sheets with their edges hosting a catalytic site. SARS-CoV-2 NendoU monomers assemble into a double-ring hexamer that is essential for enzymatic activity.
The active site is located in a shallow groove between the two ꞵ-sheets and contains six key residues: His235, His250, Lys290, Thr341, Tyr343, and Ser294. These key residues are conserved among SARS-CoV-2, SARS-CoV, and MERS-CoV. However, the MERS-CoV enzyme shows significant structural changes, particularly in the middle domain, that could impact enzymatic activity. Specifically, key residues postulated to be responsible for metal binding in SARS-CoV-2 and SARS-CoV are absent in the case of MERS-CoV. This key difference would alter the catalytic behavior of MERS-CoV in comparison to SARS-CoV-2 and SARS-CoV. Since many therapeutics are designed to target the enzymatic active site of a virus, Kim et al. postulate that inhibitors designed for SARS-CoV-2 would cross-react with SARS-CoV but would be unlikely to inhibit the enzyme in MERS-CoV.
The structural insights gained through this work significantly increases the understanding of the structure and function of NendoU in coronaviruses. And the results highlight specific variations in virus enzyme active site that could lead to the development of custom therapeutics capable of specifically targeting the key replication engine of the virus. ― Stephen Taylor
See: Youngchang Kim1,2, Robert Jedrzejczak1,2, Natalia I. Maltseva1,2, Mateusz Wilamowski1, Michael Endres2, Adam Godzik1, Karolina Michalska1,2, and Andrzej Joachimiak1,2*, “Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2,” Prot. Sci. 29, 1596 (2020). DOI: 10.1002/pro.3873
Author affiliations: 1The University of Chicago, 2Argonne National Laboratory, 3University of California Riverside
Correspondence: * andrzejj@anl.gov
We thank the members of SBC-XSD at Argonne National Laboratory, especially Darren Sherrell and Alex Lavens for their help with setting beamline and data collection. Funding for this project was provided in part by federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract HHSN272201700060C. The use of SBC-XSD beamlines at the Advanced Photon Source is supported by the U.S. Department of Energy (DOE) Office of Science and operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DEAC02-06CH11357. This research used resources of the Advanced Photon Source, a U.S. DOE Office of Science User Facility, operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. Extraordinary facility operations were supported in part by the DOE Office of Science through the National Virtual Biotechnology Laboratory, a consortium of DOE national laboratories focused on the response to COVID-19, with funding provided by the Coronavirus CARES Act.
The U.S. Department of Energy's APS is one of the world’s most productive x-ray light source facilities. Each year, the APS provides high-brightness x-ray beams to a diverse community of more than 5,000 researchers in materials science, chemistry, condensed matter physics, the life and environmental sciences, and applied research. Researchers using the APS produce over 2,000 publications each year detailing impactful discoveries, and solve more vital biological protein structures than users of any other x-ray light source research facility. APS x-rays are ideally suited for explorations of materials and biological structures; elemental distribution; chemical, magnetic, electronic states; and a wide range of technologically important engineering systems from batteries to fuel injector sprays, all of which are the foundations of our nation’s economic, technological, and physical well-being.
Argonne National Laboratory seeks solutions to pressing national problems in science and technology. The nation's first national laboratory, Argonne conducts leading-edge basic and applied scientific research in virtually every scientific discipline. Argonne researchers work closely with researchers from hundreds of companies, universities, and federal, state and municipal agencies to help them solve their specific problems, advance America's scientific leadership and prepare the nation for a better future. With employees from more than 60 nations, Argonne is managed by UChicago Argonne, LLC, for the U.S. DOE Office of Science.
The U.S. Department of Energy's Office of Science is the single largest supporter of basic research in the physical sciences in the United States and is working to address some of the most pressing challenges of our time. For more information, visit the Office of Science website.
