It is well known that DNA is critical to life itself, playing a role in almost everything we do and impacting our health as well as disease. But what happens when this crucial but microscopically-thin double-stranded helix of nucleotides breaks? Researchers seeking to learn more about the answer to this question published results in Nature Communications based on work conducted at the U.S. Department of Energy’s Advanced Photon Source (APS). The work involved generating crystals that captured pre-catalytic and catalytic snapshots, in crystallo, of a DNA repair enzyme, Polµ, at the site of a DNA break, providing insights into the role of Polµ in DNA repair that will enhance our understanding of cancer and other diseases in which DNA breaks may play a role.
DNA can be broken by outside factors, such as ionizing radiation, or internal factors, such as reactive oxygen species that are generated through normal cellular metabolism, resulting in single-strand breaks (SSBs) or double-strand breaks (DSBs) in the DNA double helix. Left unrepaired, these DNA breaks can lead to cell death or diseases, like cancer. Of course, given the importance of genomic integrity, cells have evolved highly efficient repair enzymes to fix DNA damage. In humans, this process involves a complex of proteins that work together to identify a break, stabilize the ends, fill in any damaged sequence, and then rejoin the ends. A key enzyme in this process is a DNA polymerase called Polµ that fills in the small sequence gaps at the broken ends to repair the break and prepare the ends for rejoining by another enzyme in the complex. Polµ belongs to a family of polymerases that engage in such work and their action on SSBs is well understood. However, questions remain regarding how these enzymes catalyze gap-filling and stabilization in DSBs and whether these reactions are similar to those carried out on SSBs.
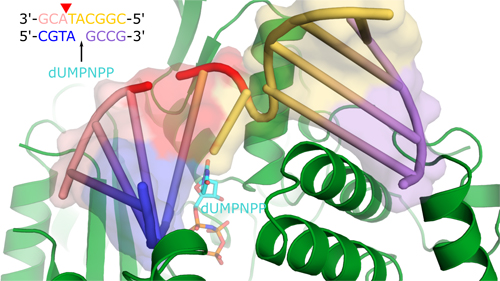
The international team of researchers started with the catalytic domain of Polµ that has the same characteristics as the full-length enzyme and crystallized it in the presence of a small piece of DNA with a double-stranded break to provide an appropriate substrate for repair (Fig. 1). In addition, they used a trick to capture “snapshots” of the enzyme in “action” in crystallo. First, they used a correctly paired non-hydrolyzable nucleotide analog to trap the structure in a pre-catalytic state and then, once they had a snapshot structure of that state, they soaked similar crystals in a catalytic solution with hydrolyzable nucleotide to capture different states during and after catalysis.
The structure of the pre-catalytic enzyme, obtained with high-brightness x-rays at the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the APS, shows that when Polµ is poised for catalysis, it stabilizes both ends of the DNA break in the substrate binding cleft with a series of van der Waals interactions, hydrogen bonds, and ionic interactions along the length of the DNA around the break and also aligns a critical upstream hydroxyl group (3’-OH) with key magnesium (Mg2+) ions required for catalysis. Once the crystal is transferred to a solution that allows the reaction to occur, the structure doesn’t shift significantly except for subtle movements around the newly forming bond, and disappearance of the pyrophosphate leaving group from the reaction, suggesting that the pre-catalytic snapshot represents a catalytically competent conformation.
Perhaps most interesting, comparison of these structures with others from this family of enzymes in complex with SSBs suggests that the enzymes perform reactions on both SSBs and DSBs in a very similar way. Although the overall conformation of the protein/DNA complex is similar, minor differences in protein DNA interactions exist between the two substrates. The rigidity of the enzyme combined with minor adjustments at the break allow Polµ to contend with a broader array of breaks than other polymerases to fill in and repair DNA. ― Sandy Field
See: Andrea M. Kaminski1, John M. Pryor2, Dale A. Ramsden2, Thomas A. Kunkel1, Lars C. Pedersen1*, and Katarzyna Bebenek1, “Structural snapshots of human DNA polymerase μ engaged on a DNA double-strand break,” Nat. Commun. 11, 4784 (2020). DOI: 10.1038/s41467-020-18506-5
Author affiliations: 1National Institutes of Health, 2University of North Carolina at Chapel Hill
Correspondence: * pederse2@niehs.nih.gov
This work was supported in part by the Division of Intramural Research of the National Institute of Environmental Health Sciences, National Institutes of Health Grants 1ZIA ES102645 (to L.C.P.) and Z01 ES065070 (to T. A.K.), and National Cancer Institute grant CA097096 (to D.A.R). SER-CAT is supported by its member institutions, and equipment grants (S10_RR25528 and S10_RR028976) from the National Institutes of Health. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
The U.S. Department of Energy's APS is one of the world’s most productive x-ray light source facilities. Each year, the APS provides high-brightness x-ray beams to a diverse community of more than 5,000 researchers in materials science, chemistry, condensed matter physics, the life and environmental sciences, and applied research. Researchers using the APS produce over 2,000 publications each year detailing impactful discoveries, and solve more vital biological protein structures than users of any other x-ray light source research facility. APS x-rays are ideally suited for explorations of materials and biological structures; elemental distribution; chemical, magnetic, electronic states; and a wide range of technologically important engineering systems from batteries to fuel injector sprays, all of which are the foundations of our nation’s economic, technological, and physical well-being.
Argonne National Laboratory seeks solutions to pressing national problems in science and technology. The nation's first national laboratory, Argonne conducts leading-edge basic and applied scientific research in virtually every scientific discipline. Argonne researchers work closely with researchers from hundreds of companies, universities, and federal, state and municipal agencies to help them solve their specific problems, advance America's scientific leadership and prepare the nation for a better future. With employees from more than 60 nations, Argonne is managed by UChicago Argonne, LLC, for the U.S. DOE Office of Science.
The U.S. Department of Energy's Office of Science is the single largest supporter of basic research in the physical sciences in the United States and is working to address some of the most pressing challenges of our time. For more information, visit the Office of Science website.
