COVID-19 has claimed millions of lives since the outbreak began in late 2019, prompting a wave of research into vaccines and therapeutics to end the pandemic. Studying the naturally occurring human antibodies created in response to SARS-CoV-2, the virus that causes COVID-19, helped researchers develop new therapies that can prevent and treat COVID-19 infections. In a study, which involved collecting x-ray diffraction data at the U.S. Department of Energy’s Advanced Photon Source (APS), researchers discovered how different antibodies bind to the SARS-CoV-2 virus and identified clues for ways to optimize antibodies so that they are more effective at targeting the virus.
When a person is exposed to a virus or given a vaccine, their immune system reacts by combining proteins from different genes and connecting these proteins to create a variety of antibodies that target a particular virus. These are called “neutralizing antibodies.” Researchers have isolated many different neutralizing antibodies from people who had been infected with COVID-19, and these antibodies have shown promise as a potential treatment for COVID-19 infections. If researchers learn more about how these neutralizing antibodies work, they may be able to use this information to develop more successful treatments or vaccines.
Neutralizing antibodies for the SARS-CoV-2 virus target a part of the virus called the “spike protein.” The spike protein helps infect host cells by interacting with angiotensin-converting enzyme 2 (ACE2) receptors on the surface of host cells. The binding between the spike protein and the ACE2 receptor allows the virus to enter the host cell. Antibodies block infection by binding to the spike protein, preventing it from being able to bind to ACE2.
A team of researchers from Columbia University, the Columbia University Vagelos College of Physicians and Surgeons, and the National Institutes of Health used various techniques to compare and contrast how different SARS-CoV-2 antibodies interact with the spike protein at a very detailed level.
First, the researchers collected information about 158 spike-specific antibodies from 10 published studies. They found that a particular class of these antibodies – those that derive from the VH1-2 antibody heavy chain gene – are especially potent. VH1-2 antibodies are known to bind to the receptor-binding domain (RBD) on the spike protein—a part of the spike protein that binds to the ACE2 receptor. The researchers concentrated their study on three different VH1-2-derived antibodies: 2-15, 2-43, and H4.
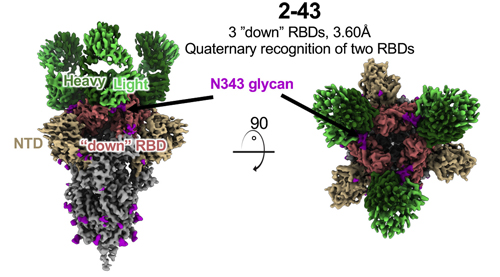
Next, the researchers used the single-particle cryo-electron microscopy (cryo-EM) technique at the National Center for Cryo-EM Access and Training and the Simons Electron Microscopy Center located at the New York Structural Biology Center, and the Columbia University Cryo-Electron Microscopy Center to create high-resolution three-dimensional reconstructions of how each antibody interacts with the spike protein. The team found differences between how the different antibodies interacted with the spike protein; While all three antibodies showed binding to the spike protein RBD, the 2-43 antibodies only bound to RBDs in a downward-facing conformation, and the H4 antibodies only bound to RBDS in an upward-facing conformation. The 2-15 antibody particles bound to a mix of upward-facing and downward-ward facing RBDs.
To get higher-resolution information about how the different antibody particles interact with the spike protein, the team used x-ray crystallography to create even more detailed three-dimensional structures of the interactions between the different antibodies and isolated spike protein fragments containing the RBD. These data were collected at the Northeastern Collaborative Access Team beamline 24-ID-C at APS (the APS is an Office of Science user facility at Argonne National Laboratory). Using this technique, the researchers were able to get more accurate information about the similarities and differences between how specific parts of each antibody interface with the RBD.
Next, the researchers compared the ways six different antibodies from the VH1-2 antibody class interacted with the RBD. The team found that all six antibodies had similar interactions with the RBDs and these interactions differed from antibodies from other antibody classes. This detailed information about how VH1-2 antibodies interact with the RBD show that different humans respond similarly to SARS-CoV-2 in their derivation of highly similar antibodies, and may help explain how these antibodies are so effective at neutralizing SARS-CoV-2 and could help determine which virus variants might be effectively neutralized—or not neutralized—by this class of antibodies. Two mutations - E484K/Q and L452R - carried by variant B.1.351 (South Africa), P.1 (Brazil) or B.1.617 (India), are within the VH1-2 antibody epitopes, and ablate the binding of this class of antibodies. The enrichment of such mutations in different emerging strains, especially for E484K/Q, suggest convergent evolution of SARS-CoV-2 by selection pressure from VH1-2 class antibodies.
The researchers then wanted to see if they could improve how well VH1-2 antibodies bind to the SARS-CoV-2 virus. They inserted a targeted mutation in both the 2-43 and 2-4 antibodies. This mutation improved how tightly both antibodies bound to the spike protein, and the mutated antibodies were also more effective at neutralizing SARS-CoV-2 virus exposed to host cells in a dish.
The results from this study provide more information about how antibodies bind to and neutralize the SARS-CoV-2 virus. They also provide clues for how to further optimize antibodies and vaccines against the SARS-CoV-2 virus, which may prove particularly important as new SARS-CoV2 variants emerge. ― Summer Allen
See: Micah Rapp1, Yicheng Guo1, 2, Eswar R. Reddem1, Jian Yu2, Lihong Liu2, Pengfei Wang2, Gabriele Cerutti1, Phinikoula Katsamba1, Jude S. Bimela1, Fabiana A. Bahna1, Seetha M. Mannepalli1, Baoshan Zhang3, Peter D. Kwong1, 3, Yaoxing Huang2, David D. Ho2, Lawrence Shapiro1, 2, 3*, and Zizhang Sheng1, 2**, “Modular basis for potent SARS-CoV-2 neutralization by a prevalent VH1-2-derived antibody class,” Cell Rep. 35, 108950 (April 6, 2021). DOI: 10.1016/j.celrep.2021.108950
Author affiliations: 1Columbia University, 2Columbia University Vagelos College of Physicians and Surgeons, 3National Institutes of Health
Correspondence: * lss8@columbia.edu, ** zs2248@cumc.columbia.edu
The National Center for Cryo-EM Access and Training and the Simons Electron Microscopy Center located at the New York Structural Biology Center is supported by the National Institutes of Health (NIH) Common Fund Transformative High Resolution Cryo-Electron Microscopy program (U24 GM129539) and by grants from the Simons Foundation (SF349247) and the NY State Assembly. Data analysis was performed at the National Resource for Automated Molecular Microscopy (NRAMM), supported by the NIH National Institute of General Medical Sciences (GM103310). We thank D. Neau, S. Banerjee, and Igor Kourinov for help with synchrotron data collection conducted at the Northeastern Collaborative Access Team 24-ID-C beamline, which is funded by the National Institute of General Medical Sciences from the NIH (P30 GM124165). Support for this work was provided by the Intramural Research Program of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases (NIAID), NIH. This study was supported by Samuel Yin, Pony Ma, Peggy, Andrew Cherng, Brii Biosciences, Jack Ma Foundation, JBP Foundation, Carol Ludwig, and Roger and David Wu. This research used resources of the APS, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357.
The U.S. Department of Energy's APS is one of the world’s most productive x-ray light source facilities. Each year, the APS provides high-brightness x-ray beams to a diverse community of more than 5,000 researchers in materials science, chemistry, condensed matter physics, the life and environmental sciences, and applied research. Researchers using the APS produce over 2,000 publications each year detailing impactful discoveries, and solve more vital biological protein structures than users of any other x-ray light source research facility. APS x-rays are ideally suited for explorations of materials and biological structures; elemental distribution; chemical, magnetic, electronic states; and a wide range of technologically important engineering systems from batteries to fuel injector sprays, all of which are the foundations of our nation’s economic, technological, and physical well-being.
Argonne National Laboratory seeks solutions to pressing national problems in science and technology. The nation's first national laboratory, Argonne conducts leading-edge basic and applied scientific research in virtually every scientific discipline. Argonne researchers work closely with researchers from hundreds of companies, universities, and federal, state and municipal agencies to help them solve their specific problems, advance America's scientific leadership and prepare the nation for a better future. With employees from more than 60 nations, Argonne is managed by UChicago Argonne, LLC, for the U.S. DOE Office of Science.
The U.S. Department of Energy's Office of Science is the single largest supporter of basic research in the physical sciences in the United States and is working to address some of the most pressing challenges of our time. For more information, visit the Office of Science website.
