The globally debilitating COVID-19 pandemic is caused by severe acute respiratory syndrome coronovirus 2 (SARS-CoV-2), which is a single-stranded RNA virus that can encode for at least 29 different proteins. One of these proteins, an enzyme named PLpro, is responsible for releasing other proteins from a larger protein complex and it plays an important function in disrupting host response. A recent high-impact paper based on research at the U.S. Department of Energy’s Advanced Photon Source (APS) and published in Nature Communications reports a high-resolution structure of the protein PLpro and provides in vitro evidence that a set of chemical compounds are capable of inhibiting the activity of this protein. Some of these compounds also demonstrate an ability to block SARS-CoV-2 from replicating in cells. Although such inhibitors will need to undergo in vivo testing in animal models before use in human clinical trials, the data presented in this work show clinical promise.
Recent studies show that the SARS-CoV-2 virus is less than 200 nanometers in diameter, has four structural proteins, three which are embedded in the lipid envelope (envelope (E), membrane (M), and spike (S)) and one that packages the RNA genome inside the viral capsid. In addition to these structural proteins, there are 15 nonstructural proteins (Nsp) and several other proteins that the SARS-CoV-2 genome encodes. One of these proteins is PLpro, a domain within Nsp3. It is an enzyme referred to as a protease. Protease enzymes cleave specific sites within proteins to produce smaller components such as individual building blocks of proteins (i.e., amino acids) or protein subunits (i.e., polypeptides). PLpro is found in all corona viruses, not just SARS-CoV-2, and cleaves a larger protein complex to liberate the three proteins Nsp1, Nsp2, and Nsp3. The protein Nsp2 has been proposed to influence the survival of infected host cells, suggesting that PLpro may be important for replication efficiency.
In a study that employed x-ray diffraction at the Structural Biology Center (SBC-XSD) 19-ID x-ray beamline, members of SBC-XSD together with researchers from The University of Chicago who are members of the National Institutes of Health/ National Institute of Allergy and Infectious Diseases-funded Center for Structural Genomics of Infectious Diseases collaborated with colleagues from the laboratories of Bryan Dickinson, Scott Snyder, and Glenn Randall (all University of Chicago) to generate high-quality crystal structures of PLpro and its complexes with inhibitors. The authors report that PLpro has a structure similar to the PLpro enzymes of SARS and Middle East Respiratory Syndrome (MERS) corona viruses. The structure shows fine molecular details of this enzyme that reveal how it works and how its action might be inhibited, which could potentially disrupt coronavirus replication. (The APS is a DOE Office of Science user facility at Argonne National Laboratory.)
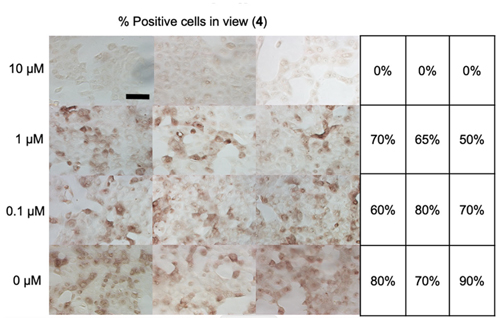
Having solved this structure, the authors then tested various compounds that inhibit PLpro for their ability to affect replication efficiency in infected cells. They found that many of these compounds inhibit the activity of this enzyme in vitro and effectively block the replication of the SARS-CoV-2 virus in infected cells. The researchers carried out their viral replication assays at the University of Chicago Ricketts Laboratory for. Ultimately, five compounds were observed to both 1) inhibit viral replication and 2) impact cell viability. Compound 4 was especially effective, as shown in Fig. 1.
Given the severity of the COVID-19 pandemic, these data are timely and tangibly meaningful. The authors have not only solved the structure of a key viral enzyme, but have provided compelling evidence that several different compounds are capable of inhibiting this protein and combating viral replication in infected cells. If these compounds are validated in pre-clinical animal models, they will be ready for human testing and could serve as novel anti-viral medications that increase the survival of patients infected with not just SARS-CoV-2, but other coronoviruses too. — Alicia Surrao
See: Jerzy Osipiuk1,2, Saara-Anne Azizi1, Steve Dvorkin1, Michael Endres1,2, Robert Jedrzejczak1,2, Krysten A. Jones1, Soowon Kang1, Rahul S. Kathayat1, Youngchang Kim1,2, Vladislav G. Lisnyak1, Samantha L. Maki1, Vlad Nicolaescu1, Cooper A. Taylor1, Christine Tesar1,2, Yu-An Zhang1, Zhiyao Zhou1, Glenn Randall1, Karolina Michalska1,2, Scott A. Snyder1, Bryan C. Dickinson1, and Andrzej Joachimiak1,2, “Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors,” Nat. Commun. 12, 743 (2021). DOI: 10.1038/s41467-021-21060-3
Author affiliations: 1The University of Chicago, 2Argonne National Laboratory
Correspondence: * sasnyder@uchicago.edu, ** dickinson@uchicago.edu, *** andrzejj@anl.gov
We thank the members of the SBC-XSD at Argonne National Laboratory, especially Darren Sherrell and Alex Lavens for their help with setting up beamline and data collection at beamline 19-ID. Funding for this project was provided in part by federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract HHSN272201700060C (to A.J.) and by the U.S. Department of Energy (DOE) Office of Science through the National Virtual Biotechnology Laboratory, a consortium of DOE national laboratories focused on response to COVID- 19, with funding provided by the Coronavirus CARES Act (to A.J.). The use of SBC-XSD beamlines at the Advanced Photon Source is supported by the U.S. DOE Office of Science and operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. Funding for the synthesis and biochemical studies was provided by a “BIG” Award from The University of Chicago, The University of Chicago Women’s Board, the National Institutes of Health (TM GM08720, Predoctoral Training Program in Chemistry and Biology, graduate fellowship to C.A.T.), the National Institute of General Medical Sciences (R35 GM119840 to B.C.D.), and start-up funds from The University of Chicago (S.A.S.). This research used resources of the Advanced Photon Source, a U.S. DOE Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. Extraordinary facility operations were supported in part by the DOE Office of Science through the National Virtual Biotechnology Laboratory, a consortium of DOE national laboratories focused on the response to COVID-19, with funding provided by the Coronavirus CARES Act.
The U.S. Department of Energy's APS is one of the world’s most productive x-ray light source facilities. Each year, the APS provides high-brightness x-ray beams to a diverse community of more than 5,000 researchers in materials science, chemistry, condensed matter physics, the life and environmental sciences, and applied research. Researchers using the APS produce over 2,000 publications each year detailing impactful discoveries, and solve more vital biological protein structures than users of any other x-ray light source research facility. APS x-rays are ideally suited for explorations of materials and biological structures; elemental distribution; chemical, magnetic, electronic states; and a wide range of technologically important engineering systems from batteries to fuel injector sprays, all of which are the foundations of our nation’s economic, technological, and physical well-being.
Argonne National Laboratory seeks solutions to pressing national problems in science and technology. The nation's first national laboratory, Argonne conducts leading-edge basic and applied scientific research in virtually every scientific discipline. Argonne researchers work closely with researchers from hundreds of companies, universities, and federal, state and municipal agencies to help them solve their specific problems, advance America's scientific leadership and prepare the nation for a better future. With employees from more than 60 nations, Argonne is managed by UChicago Argonne, LLC, for the U.S. DOE Office of Science.
The U.S. Department of Energy's Office of Science is the single largest supporter of basic research in the physical sciences in the United States and is working to address some of the most pressing challenges of our time. For more information, visit the Office of Science website.
