Finding peptides that bind to proteins of interest — such as those that play key roles in human diseases and disorders — can be expensive and time-consuming. In the past few decades, this process has been greatly facilitated through phage display. This technique involves engineering the gene for various peptides in frame with the phage coat protein, prompting the phage, a virus that can infect and kill bacteria, to display the peptide on its surface. Although this method has led to the discovery of hundreds of potential target-specific hits, these hits must still be synthesized to confirm binding as a free peptide, which adds considerable effort and cost and has low yield. Furthermore, to identify peptides that bind to specific regions of interest on the target protein, researchers have traditionally used a method called alanine mutation, which substitutes alanine for other individual amino acids at a particular site to change binding propensity. However, the process of cloning, expressing, purifying, and characterizing each alanine mutation is also laborious and time consuming. To get around these challenges and smooth the process of discovering functional peptides, users of the U.S. Department of Energy’s Advanced Photon Source (APS) tried a new tactic: They combined phage display with a related platform called yeast display, incorporating target proteins and their alanine-mutated variants into the cell walls of yeast. The authors of this study, published in the journal PLOS ONE, suggest that this approach could be applied to any target protein that can be displayed on yeast, narrows the number of proteins that ultimately require synthesis, and enriches the population of promising target-specific peptides, providing a reliable and cost-effective way to improve peptide discovery.
As a model target protein, the researchers selected interleukin 23 (IL-23), a chemical messenger that plays a key role in several autoimmune diseases. Although IL-23 is composed of multiple subunits, they were particularly interested in one known as p19, which provides a unique binding area that’s not shared by other interleukin 12.
The team created peptide libraries of different lengths against IL-23 — including cyclic 12, 15, and 18 libraries of random amino acids, as well as one linear library 15 amino acids long — displayed on phages. They also created yeast that displayed wild-type IL-23 or alanine-mutated variants of the p19 subunit in complex with p40 on their surfaces.
To validate whether a phage-displayed peptide could bind to a yeast-displayed target, the researchers tested this concept with phage that displayed a key binding region of an antibody (Fab) that had already been discovered to bind to the p19 region of IL-23. Although a naked phage didn’t bind to the yeast displaying wild-type IL-23 or any of its alanine mutants, the one displaying part of the previously discovered antibody bound to nearly all of these, with the exception of yeast carrying a mutation in a residue that’s known to be a major site for receptor binding.
With evidence that this approach is feasible, the researchers tested the binding of the peptides as displayed on phage to wild-type and select alanine mutants of p19 displayed on yeast. Consequently, they identified several different phage-peptides in their library that bound strongly to yeast displaying wild-type IL-23 and not to the p19 alanine mutants that correspond to the peptide binding site. Using a technique called hydrogen deuterium exchange mass spectrometry, they confirmed p19-specific binding: While the p40 subunit of wild-type IL-23 had significant deuterium uptake when it was exposed to this hydrogen isotope, suggesting that it remained open and exposed, the p19 subunit didn’t take up much deuterium, suggesting that its binding to these peptides blocked deuterium exchange.
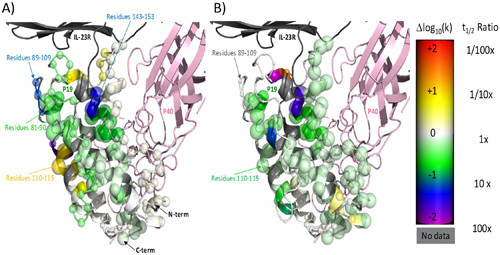
To further investigate binding, the researchers used the Lilly Research Laboratories Collaborative Access Team (LRL-CAT) 31-ID-D beamline at the APS to perform x-ray crystallography, which allowed them to see crystal structures of these phage peptides bound and unbound to IL-23 (Fig. 1), providing additional confirmation that these peptides bound to the p19 subunit. (The APS is an Office of Science user facility at Argonne National Laboratory.)
To provide even more confirmation that these phage-displayed peptides targeted IL-23, and specifically p19, the researchers synthesized free peptides of four of the most promising phage-displayed peptides and showed that these could successfully compete with IL-23 receptors to bind to IL-23 proteins. Additional protein-protein interaction assays, known as AlphaLISA and TR-FRET, further showed that these four peptides blocked IL-23 from binding to its receptor.
Finally, the researchers showed that this whole process of identifying promising phage-displayed peptides using yeast-displayed proteins could be accelerated using a cell sorting technique. ― Christen Brownlee
See: Priyanka Pandya1, Robert O. Sayers1, Joey P. Ting1, Shaghayegh Morshedian1, Carina Torres1, Justine S. Cudal1, Kai Zhang1, Jonathan R. Fitchett1, Qing Zhang1, Feiyu F. Zhang2, Jing Wang2, Jim D. Durbin3, Juan J. Carrillo3, Alfonso Espada4, Howard Broughton4, Yuewei Qian2, and Sepideh Afshar1*, “Integration of phage and yeast display platforms: A reliable and cost effective approach for binning of peptides as displayed on-phage,” PLoS ONE 15(6), e0233961 (June 1 2020). DOI: 10.1371/journal.pone.0233961
Author affiliations: 1Eli Lilly Biotechnology Center, 2Lilly Research Laboratories, 3Eli Lilly and Company, 4Centro de Investigación Lilly
Correspondence: *s.afshar@lilly.com
Use of the Lilly Research Laboratories Collaborative Access Team (LRL-CAT) beamline at Sector 31 of the Advanced Photon Source was provided by Eli Lilly Company, which operates the facility. Use of the Advanced Photon Source, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the U.S. DOE under Contract No. DE-AC02-06CH11357.
The U.S. Department of Energy's APS is one of the world’s most productive x-ray light source facilities. Each year, the APS provides high-brightness x-ray beams to a diverse community of more than 5,000 researchers in materials science, chemistry, condensed matter physics, the life and environmental sciences, and applied research. Researchers using the APS produce over 2,000 publications each year detailing impactful discoveries, and solve more vital biological protein structures than users of any other x-ray light source research facility. APS x-rays are ideally suited for explorations of materials and biological structures; elemental distribution; chemical, magnetic, electronic states; and a wide range of technologically important engineering systems from batteries to fuel injector sprays, all of which are the foundations of our nation’s economic, technological, and physical well-being.
Argonne National Laboratory seeks solutions to pressing national problems in science and technology. The nation's first national laboratory, Argonne conducts leading-edge basic and applied scientific research in virtually every scientific discipline. Argonne researchers work closely with researchers from hundreds of companies, universities, and federal, state and municipal agencies to help them solve their specific problems, advance America's scientific leadership and prepare the nation for a better future. With employees from more than 60 nations, Argonne is managed by UChicago Argonne, LLC, for the U.S. DOE Office of Science.
The U.S. Department of Energy's Office of Science is the single largest supporter of basic research in the physical sciences in the United States and is working to address some of the most pressing challenges of our time. For more information, visit the Office of Science website.
