Mithramycin (MTM) is an anticancer antibiotic produced by the gram-positive bacterium Streptomyces argillaceus. The enzymes MtmOIV and MtmW were found to catalyze the final two reactions in mithramycin’s biosynthetic pathway, the Baeyer–Villiger opening of the fourth ring of premithramycin B (PMB), creating a C3 pentyl side chain, followed closely by reduction of the distal keto group on the new side chain. Unexpectedly, the completion of this biosynthetic pathway yields only a small amount of active MTM and a large amount of an inert form of MTM, dubbed iso‐MTM. Iso-MTM was then observed to non-enzymatically convert to the toxic MTM species in the presence of magnesium ions. The reason for this roundabout series of production steps is presumably a previously unobserved form of self-resistance to MTM, which is toxic to gram-positive bacteria. X-ray crystallography carried out at the U.S. Department of Energy’s Advanced Photon Source to examine the high-resolution structures of MtmW in complex with the co‐substrate NADP+ and polyethylene glycol suggests a catalytic mechanism for MtmW. The results and structures, published in the journal Angewandte Chemie International Edition also show that the assembly of MtmW closely resembles the ring‐shaped β subunit of a vertebrate ion channel, suggesting the enzyme may play a role in exporting iso-MTM out of the cell prior to its activation to a toxic agent. This unusual transporter-mediated mechanism may shed light on the production of other potentially clinically useful natural products.
Small-molecule natural products, which have evolved structurally to exert specific biological functions over millions of years, provide the producer organisms with evolutionary advantages. As chemical warfare agents, these molecules usually bind house-keeping elements of the competing organisms and inhibit their normal life cycles. Such natural products, however, may create similar threats to the producing organisms themselves. To avoid self-destruction, many natural-product biosynthetic gene clusters encode not only enzymes for their production, but also additional regulatory factors to control the expression of the genes, transporters to ensure the toxic molecule is efficiently expelled from the producer organism, or dedicated resistance mechanisms, such as enzymes that inactivate the product molecules that linger in the cytoplasm.
The producer of the anticancer antibiotic mithramycin (MTM), the bacterium Streptomyces argillaceus, appears to have evolved a rather intriguing form of protection against such self-destruction. Besides being an antibacterial agent, MTM is a potent transcriptional inhibitor targeting G/C-rich promoters that has been found to inhibit growth of cancer cells dependent on oncogenic transcription. Specifically, MTM is a highly potent antagonist of the bone and soft tissue cancer Ewing sarcoma and prostate cancers that depend on similar oncogenic transcription mechanisms.
To gain a better understanding of the final biosynthetic steps of mithramycin for the development of potential drugs to treat Ewing sarcoma and prostate cancers, researchers from the University of Kentucky and the University of Texas Health Science Center focused on the enzymes responsible for the generation of mithramycin’s C3-side chain. They discovered that the enzymatic cooperation of the enzymes MtmOIV (a Baeyer–Villiger monooxygenase) and MtmW (a ketoreductase) is essential for MTM formation, through the creation of a transient complex.
Surprisingly, the researchers discovered that this biosynthetic pathway actually produces scant amounts of MTM and large amounts of a stereoisomer of MTM that is biologically inert. This inert form, called iso-MTM, is converted to MTM in the presence of environmental magnesium ions, presumably after the compound has been expelled from the organism that produced it.
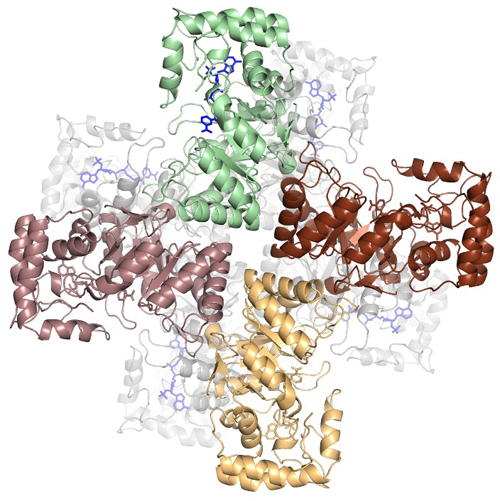
The research team then used x-ray diffraction data collected on the Southeast Regional Collaborative Access Team synchrotron beamline 22-ID at the APS (the APS is a DOE Office of Science user facility at Argonne National Laboratory) to determine the crystal structures of MtmW in two forms, with the apo-MtmW crystallized in one form (diffracting to 1.8 Å resolution) and its complex with NADP+ in the other form (2.1 angstrom resolution, Fig. 1). Additionally, the similarity of the crystal structures of MtmW to a ring‐shaped β subunit of a vertebrate ion channel Kvb indicated that the activity of MtmW may be coupled to channeling mithramycin out of the organism through the transmembrane channel MtrAB.
The formation and export of an inert precursor (iso-MTM) would explain the self-resistance of the producing organism to the toxic MTM. This somewhat unusual self-resistance mechanism may be used by Streptomyces argillaceus and other bacteria. Knowledge of this mechanism may help in the discovery and utilization of other natural products with important uses. ― Chris Palmer
See: Ryan Wheeler1, Xia Yu1,2, Caixia Hou1, Prithiba Mitra1, Jhong-Min Chen1, Frank Herkules3, Dmitri N. Ivanov3, Oleg V. Tsodikov1*, and Jgrgen Rohr1**, “Discovery of a Cryptic Intermediate in Late Steps of Mithramycin Biosynthesis,” Angew. Chem. Int. Ed. 59, 826 (2020). DOI: 10.1002/anie.201910241
Author affiliations: 1University of Kentucky, 2Central South University Changsha, 3University of Texas Health Science Center
Correspondence: *oleg.tsodikov@uky.edu, **jrohr2@uky.edu
The work was supported by National Institutes of Health (NIH) grants CA 091901 and GM 1051977 to J.R. The Southeast Regional Collaborative Access Team is supported by its member institutions (see www.ser-cat.org/members.html), and equipment grants (S10_RR25528, S10_RR028976 and S10_OD027000) from the NIH. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
The U.S. Department of Energy's APS is one of the world’s most productive x-ray light source facilities. Each year, the APS provides high-brightness x-ray beams to a diverse community of more than 5,000 researchers in materials science, chemistry, condensed matter physics, the life and environmental sciences, and applied research. Researchers using the APS produce over 2,000 publications each year detailing impactful discoveries, and solve more vital biological protein structures than users of any other x-ray light source research facility. APS x-rays are ideally suited for explorations of materials and biological structures; elemental distribution; chemical, magnetic, electronic states; and a wide range of technologically important engineering systems from batteries to fuel injector sprays, all of which are the foundations of our nation’s economic, technological, and physical well-being.
Argonne National Laboratory seeks solutions to pressing national problems in science and technology. The nation's first national laboratory, Argonne conducts leading-edge basic and applied scientific research in virtually every scientific discipline. Argonne researchers work closely with researchers from hundreds of companies, universities, and federal, state and municipal agencies to help them solve their specific problems, advance America's scientific leadership and prepare the nation for a better future. With employees from more than 60 nations, Argonne is managed by U Chicago Argonne, LLC, for the U.S. DOE Office of Science.
The U.S. Department of Energy's Office of Science is the single largest supporter of basic research in the physical sciences in the United States and is working to address some of the most pressing challenges of our time. For more information, visit the Office of Science website.
