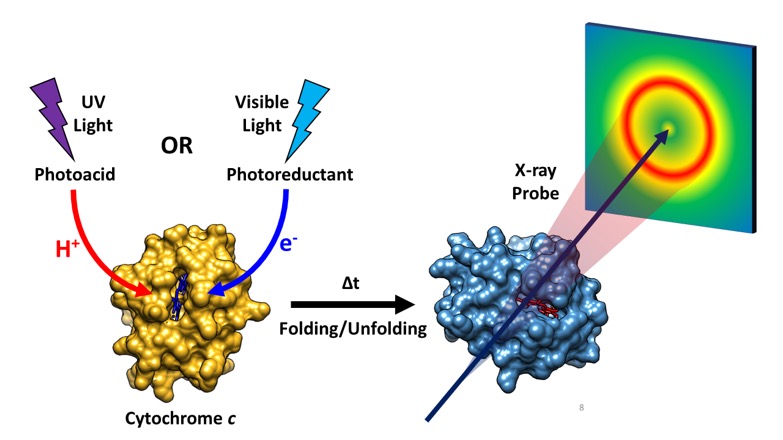
The direct tracking of protein structural dynamics during folding and unfolding processes is important for understanding the formation of functional proteins. In an effort to monitor such dynamics, a team of researchers employed time-resolved x-ray solution scattering using the U.S. Department of Energy’s Advanced Photon Source (APS) to investigate structural changes during the folding of the cytochrome c, a large transmembrane protein involved in the respiratory electron transport chain. Their results, published in the Journal of Physical Chemistry B, demonstrate how different environmental conditions can change the nature of interaction between a protein’s active site and its overall conformation, even within the same metalloprotein, which subsequently affect folding structural dynamics. The team believes that the experimental technique employed here, time-resolved x-ray solution scattering or TRXSS, coupled to jumps in pH, electron transfer, and previously reported jumps in temperature, will make it possible to perform detailed studies of biologically relevant processes in non-photoactive proteins with high structural sensitivity and high temporal resolution at the same time.
Protein structure-function relationships are known to be sensitive to environmental conditions, such as temperature, pH, and the presence of compounds that trigger redox reactions. Within cells, protein environment, structure, and function work together to regulate a protein’s biological functions via conformational gating or ligand binding. Protein folding dynamics typically span multiple timescales and can range from nanoseconds to milliseconds for fast folding proteins.
Traditional time-resolved structural biology methods, such as stopped flow or continuous flow mixing, typically cannot resolve the first 100 microseconds from initiating mixing and the beginning of kinetics observations, making these methods not suited to tracking dynamics on short timescales. In contrast, pulsed laser-based methods can initiate reactions on timescales as short as femtoseconds, making them well suited to observations of short-lived intermediate states.
Still, laser-based approaches are limited in that naturally occurring light-driven proteins are rare and an intrinsic chromophore is needed to drive the dynamics and probe structural changes. The team of researchers from Northwestern University, the University of Delaware, The University of Chicago, and Argonne National Laboratory devised a new technique for studying protein folding dynamics that eliminated the need for an intrinsic chromophore by employing laser excitation to perturb the protein environment. They then probed the resulting structural changes with TRXSS, carried out at the BioCARS 14-ID-B beamline at the APS, which is capable of directly tracking protein tertiary structural dynamics, which gives it an important advantage over spectroscopic methods such as optical transient absorption (OTA) that indirectly infers global protein conformation. The team also collected steady-state scattering patterns of proteins at different temperature at the DuPont-Northwestern-Dow Collaborative Access Team 5-ID-B,C,D beamline at the APS. These patterns served as reference in data analysis to extract transient protein structures. (The APS is an Office of Science user facility at Argonne National Laboratory.)
The protein that the researchers investigated, cytochrome c, is a heme metalloprotein that takes part in the mitochondrial electron transport chain. It also plays a role in apoptosis, or programmed cell death. In their study, the researchers triggered protein conformational changes in cytochrome c by either bombarding the protein with UV light to induce a jump in pH or with visible light to induce electron transfer. Both experiments utilized TRXSS to directly observe the resulting structural dynamics as the protein underwent folding and unfolding (Fig. 1).
Starting from partially unfolded cytochrome c, a sudden pH drop caused a structural contraction within microseconds, followed by active site restructuring and unfolding within milliseconds. In contrast, the reduction of iron in the heme embedded within cytochrome c produced by photoinduced electron transfer did not affect the protein’s conformational stability at sub-millisecond timescales, despite changes to active site coordination geometry.
The team found the TRXSS experiments to be sensitive to both the active site dynamics and the global structural rearrangements in cytochrome c. These properties of TRXSS allowed the researchers to directly observe the formation of a previously unseen intermediate state in the pH-jump experiments, which did not affect the active site dynamics directly and would therefore have been missed with OTA. — Chris Palmer
See: Dolev Rimmerman1, Denis Leshchev1, Darren J. Hsu1, Jiyun Hong1, Baxter Abraham2, Robert Henning3, Irina Kosheleva3, and Lin X. Chen1,4*, “Probing Cytochrome c Folding Transitions upon Phototriggered Environmental Perturbations Using Time-Resolved X‑ray Scattering,” J. Phys. Chem. B 122, 5218 (2018). DOI: 10.1021/acs.jpcb.8b03354
Author affiliations: 1Northwestern University, 2University of Delaware, 3The University of Chicago, 4Argonne National Laboratory
Correspondence: * l-chen@northwestern.edu, lchen@anl.gov
This work was supported by the National Institutes of Health (NIH), under contract no. R01-GM115761. B.A. acknowledges support from the U.S. Department of Energy (DOE) Office of Science Graduate Student Research program, administered by the Oak Ridge Institute for Science and Education, managed by ORAU under contract number DE-SC0014664, as well as from the U.S. DOE Office of Science-Basic Energy Sciences, under award number DE-SC0016288. Use of BioCARS was also supported by the National Institute of General Medical Sciences of the NIH under grant number R24GM111072. Time-resolved setup at Sector 14 was funded in part through collaboration with Philip Anfinrud (NIH/NIDDK). The authors also acknowledge Guy Macha (BioCARS) for his assistance in designing the sample holder. The DuPont-Northwestern-Dow Collaborative Access Team is supported by Northwestern University, E.I. DuPont de Nemours & Co., and The Dow Chemical Company. Data were collected using an instrument funded by the National Science Foundation under award number 0960140. This research used resources of the Advanced Photon Source, a U.S. DOE Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under contract no. DE-AC02-06CH11357.
