The original Harvard Medical School press release by Ekaterina Pesheva can be read here.
The human body is built for survival. Each one of its cells is closely guarded by a set of immune proteins armed with nearly foolproof radars that detect foreign or damaged DNA. One of the cells’ most critical sentinels is a “first responder” protein known as cGAS, which senses the presence of foreign and cancerous DNA and initiates a signaling cascade that triggers the body’s defenses. The 2012 discovery of cGAS ignited a firestorm of scientific inquiry, resulting in more than 500 research publications, but the structure and key features of the human form of the protein continued to elude scientists. Now, scientists at the Harvard Medical School and the Dana-Farber Cancer Institute, working at the U.S. Department of Energy’s Advanced Photon Source, have for the first time identified the structural and functional differences in human cGAS that set it apart from cGAS in other mammals and underlie its unique function in people.
A report on the team’s work, published in the journal Cell, outlines the protein’s structural features that explain why and how human cGAS senses certain types of DNA, while ignoring others.
“The structure and mechanism of action of human cGAS have been critical missing pieces in immunology and cancer biology,” said senior investigator Philip Kranzusch, assistant professor of microbiology and immunobiology at the Harvard Medical School and the Dana-Farber Cancer Institute. “Our findings detailing the molecular makeup and function of human cGAS close this critical gap in our knowledge.” Importantly, the findings can inform the design of small-molecule drugs tailored to the unique structural features of the human protein—an advance that promises to boost the precision cGAS-modulating drugs that are currently in development as cancer therapies. “Several promising experimental immune therapies currently in development are derived from the structure of mouse cGAS, which harbors key structural differences with human cGAS,” Kranzusch said. “Our discovery should help refine these experimental therapies and spark the design of new ones. It will pave the way toward structure-guided design of drugs that modulate the activity of this fundamental protein.”
The team’s findings explain a unique feature of the human protein—its capacity to be highly selective in detecting certain types of DNA and its propensity to get activated far more sparingly, compared with the cGAS protein in other animals. Specifically, the research shows that human cGAS harbors mutations that make it exquisitely sensitive to long lengths of DNA but render it “blind” or “insensitive” to short DNA fragments.
“Human cGAS is a highly discriminating protein that has evolved enhanced specificity toward DNA,” said co-first author Aaron Whiteley, a postdoctoral researcher in the Department of Microbiology and Immunobiology at Harvard Medical School. “Our experiments reveal what underlies this capability.”
In all mammals, cGAS works by detecting DNA that’s in the wrong place. Under normal conditions, DNA is tightly packed and protected in the cell’s nucleus—the cellular “safe”—where genetic information is stored. DNA has no business roaming freely around the cell. When DNA fragments do end up outside the nucleus and in the cell’s cytosol, the liquid that encases the cell’s organelles, it’s usually a sign that something ominous is afoot, such as damage coming from within the cell or foreign DNA from viruses or bacteria that has made its way into the cell.
The cGAS protein works by recognizing such misplaced DNA. Normally, it lies dormant in cells. But as soon as it senses the presence of DNA outside the nucleus, cGAS springs into action. It makes another chemical—a second messenger—called cGAMP, thus setting in motion a molecular chain reaction that alerts the cell to the abnormal presence of DNA. At the end of this signaling reaction, the cell either gets repaired or, if damaged beyond repair, it self-destructs.
But the health and integrity of the cell are predicated on cGAS’ ability to distinguish harmless DNA from foreign DNA or self-DNA released during cell damage and stress. “It’s a fine balancing act that keeps the immune system in equilibrium. An overactive cGAS can spark autoimmunity, or self-attack, while cGAS that fails to detect foreign DNA can lead to tumor growth and cancer development,” said co-first author Wen Zhou, a postdoctoral researcher at Harvard Medical School and Dana-Farber Cancer Institute.
The current study reveals the evolutionary changes to the protein’s structure that allow human cGAS to ignore some DNA encounters while responding to others.
For their work, the team turned to an unlikely collaborator—Vibrio cholerae, the bacterium that causes cholera, one of humankind’s oldest scourges.
Taking advantage of a cholera enzyme that shares similarities with cGAS, the scientists were able to recreate the function of both human and mouse cGAS in the bacterium.
Teaming up with colleagues from the lab of Harvard Medical School bacteriologist John Mekalanos, the scientists designed a chimeric, or hybrid, form of cGAS that included genetic material from both the human and mouse forms of the protein. Then they compared the ability of the hybrid cGAS to recognize DNA against both the intact mouse and intact human versions of the protein.
In a series of experiments, the scientists observed activation patterns between the different types of cGAS, progressively narrowing down the key differences that accounted for differential DNA activation among the three. The experiments revealed that out of the 116 amino acids that differ in human and mouse cGAS, only two accounted for the altered function of human cGAS. Indeed, human cGAS was capable of recognizing long DNA with great precision but it ignored short DNA fragments. The mouse version of the protein, by contrast, did not differentiate between long and short DNA fragments.
“These two tiny amino acids make a world of difference,” Whiteley said. “They allow the human protein to be highly selective and respond only to long DNA, while ignoring short DNA, essentially rendering the human protein more tolerant of DNA presence in the cytosol of the cell.”
Plotting the genetic divergence on an evolutionary timescale, the scientists determined that the human and mouse cGAS genes parted ways sometime between 10 million and 15 million years ago.
The two amino acids responsible for sensing long DNA and tolerating short DNA are found solely in humans and nonhuman primates, such as gorillas, chimps and bonobos. The scientists hypothesize that the ability to ignore short DNA but recognize long DNA must have conferred some evolutionary benefits. “It could be a way to guard against an overactive immune system and chronic inflammation,” Kranzusch said. “Or it could be that the risk of certain human diseases is lowered by not recognizing short DNA.”
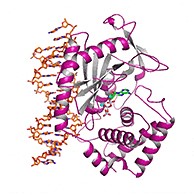
In a final set of experiments, the team determined the atomic structure of the human cGAS in its active form as it binds to DNA. To do so, they collected x-ray diffraction data at the Northeastern Collaborative Access Team 24-ID-C and 24-ID-E x-ray beamlines of the Advanced Photon Source, (the Advanced Photon Source, is an Office of Science user facility at Argonne National Laboratory).
Profiling the structure of cGAS “in action” revealed the precise molecular variations that allowed it to selectively bind to long DNA, while ignoring short DNA.
“Understanding what makes the structure and function of human cGAS different from those in other species was the missing piece,” Kranzusch said. “Now that we have it, we can really start designing drugs that work in humans, rather than mice.”
See: Wen Zhou1,2, Aaron T. Whiteley1,2, Carina C. de Oliveira Mann1,2, Benjamin R. Morehouse1,2, Rados1, P. Nowak1,2, Eric S. Fischer1,2, Nathanael S. Gray1,2, John J. Mekalanos1, and Philip J. Kranzusch1,2*, “Structure of the Human cGAS–DNA Complex Reveals Enhanced Control of Immune Surveillance,” Cell 174, 300 (July 12, 2018). DOI: 10.1016/j.cell.2018.06.026
Author affiliations: 1Harvard Medical School, 2Dana-Farber Cancer Institute
Correspondence: *philip_kranzusch@dfci.harvard.edu
The Office of Science is the single largest supporter of basic research in the physical sciences in the United States and is working to address some of the most pressing challenges of our time. For more information please visit https://science.energy.gov.
