The crystal structure for an enzyme that plays a key role in DNA methylation, the process by which methyl groups are added to the DNA molecule, has been solved by a research team led by a biochemist at the University of California, Riverside. The structure is derived from data obtained at the U.S. Department of Energy’s Advanced Photon Source (APS) and Advanced Light Source (ALS) national synchrotron x-ray research facilities. The study results were published in the journal Nature.
DNA methylation alters gene expression. This fundamental cellular mechanism critically influences plant, animal, and human development. It is known to regulate genome stability and cell differentiation. In humans, errors in methylation have been associated with various diseases, including cancer.
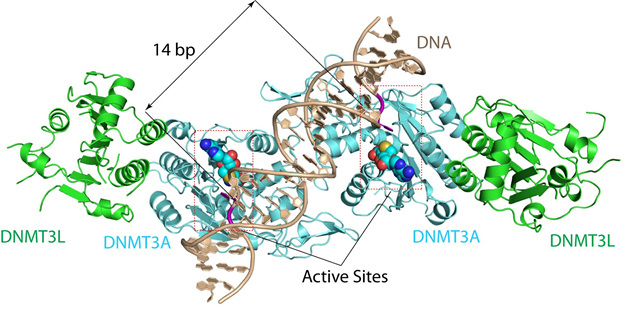
In mammals, DNA methylation is established de novo (afresh) by closely related enzymes, DNMT3A and DNMT3B, during germ cell development and early embryonic development. One difficulty in understanding how de novo DNA methylation works is that the structures of these enzymes are not known.
The UC Riverside team who led structural and biochemical studies, together with colleagues from the University of North Carolina at Chapel Hill School of Medicine who led functional studies, have now solved the crystal structure for substrate-bound DNMT3A and have further carried out functional studies in cells. X-ray diffraction datasets were obtained at the Advanced Light Source x-ray beamlines BL501 and BL502, at the Lawrence Berkeley National Laboratory, and at the Northeastern Collaborative Access Team 24-ID-E beamline at the Advanced Photon Source at Argonne National Laboratory. Both the ALS and APS are Office of Science user facilities.
This breakthrough reveals how the enzyme recognizes and methylates its substrates — important information for understanding de novo DNA methlylation. A substrate is a material or substance on which an enzyme acts.
“The structure reveals that DNMT3A molecules attack two substrate sites adjacent to each other on the same DNA molecule,” said Jikui Song, a UC Riverside associate professor of biochemistry who led the research project. “This now offers us a much clearer view on how de novo DNA methylation takes place. Our work presents the first structural view of de novo DNA methylation and presents a model for how some DNMT3A mutations contribute to cancers, such as acute myeloid leukemia. This study should provide important insights into the function of DNMT3B as well.”
Song explained the structural knowledge of DNMT3A will allow scientists to control DNA methylation content, gene expression, and cell differentiation — all of which are linked to diseases and finding cures for them.
“It especially has important implications in cancer therapy in the long term,” he said.
The DNMT3A structure that Song’s team cracked explains why mammalian DNA methylation predominantly occurs at “CpG dinucleotides” — DNA locations where cytosine nucleotides are next to guanidine nucleotides.
“Before our study, why mammalian DNA methylation mostly occurs at the CpG sites was not understood, and our understanding of de novo DNA methylation was purely based on computational modeling, which cannot reliably explain how DNMT3A works,” Song said. “Just how DNMT3A succeeded in binding to its substrate was not understood either. Our structure for DNMT3A-DNA complex addresses all these concerns, offering a far better understanding of how specific DNA methylation patterns are generated.”
The study of DNMT3A structure with substrates has long been hindered by the difficulty in producing a stable enzyme-substrate complex.
“To overcome this challenge, we successfully developed a method to trap the reaction intermediate of DNMT3A-substrate complex, and solved the structure by x-ray crystallography,” Song said.
See: Zhi-Min Zhang1‡, Rui Lu2, Pengcheng Wang1, Yang Yu1, Dongliang Chen2, Linfeng Gao1, Shuo Liu1, Debin Ji1, Scott B Rothbart2,3, Yinsheng Wang1, Gang Greg Wang2**, and Jikui Song1*, “Structural basis for DNMT3A-mediated de novo DNA methylation,” Nature, published online 07 February 2018. DOI: 10.1038/nature25477.
Author affiliations: 1University of California, Riverside, 2University of North Carolina at Chapel Hill School of Medicine, 3Van Andel Research Institute ‡Present address: Jinan University
Correspondence: *jikui.song@ucr.edu, **greg_wang@med.unc.edu
We thank staff members at the Advanced Light Source, Lawrence Berkeley National Laboratory, and at the Advanced Photo Source, Argonne National Laboratory, for access to x-ray beamlines. We are also grateful for the support of University of North Carolina facilities including Genomics Core, which are partly supported by UNC Cancer Center Core Support Grant P30-CA016086. This work was supported by Kimmel Scholar Awards (to J.S. and G.G.W.), the March of Dimes Foundation (1-FY15-345 to J.S.), the DoD Peer-reviewed Cancer Research Program (W81XWH-14-1-0232 to G.G.W.), Gabrielle’s Angel Foundation for Cancer Research (to G.G.W.), Gilead Sciences Research Scholars Program in haematology/oncology (to G.G.W.), University Cancer Research Fund of the N.C. state (to G.G.W.), and the National Institutes of Health (1R35GM119721 to J.S.; R35GM124736 to S.B.R.; 5R21ES025392 to Y.W.; and 1R01CA215284, 1R01CA218600, and 1R01CA211336 to G.G.W.). G.G.W. is a Research Scholar of American Cancer Society and a Junior Faculty Scholar of American Society of Haematology. R.L. was supported by a Lymphoma Research Foundation postdoctoral fellowship. Research conducted at the Northeastern Collaborative Access Team beamlines are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P41 GM103403). The Eiger 16M detector on the 24-ID-E beamline is funded by a NIH-ORIP HEI grant (S10OD021527. This research used resources of the Advanced Light Source, which is a DOE Office of Science User Facility under contract no. DE-AC02-05CH11231. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
Argonne National Laboratory seeks solutions to pressing national problems in science and technology. The nation's first national laboratory, Argonne conducts leading-edge basic and applied scientific research in virtually every scientific discipline. Argonne researchers work closely with researchers from hundreds of companies, universities, and federal, state and municipal agencies to help them solve their specific problems, advance America's scientific leadership and prepare the nation for a better future. With employees from more than 60 nations, Argonne is managed by UChicago Argonne, LLC for the U.S. Department of Energy's Office of Science.
The U.S. Department of Energy's Office of Science is the single largest supporter of basic research in the physical sciences in the United States and is working to address some of the most pressing challenges of our time. For more information, visit the Office of Science website.
