The Princeton University blog entry by the Staff of the Department of Molecular Biology can be read here.
Researchers from Princeton University and the National Institutes of Health have discovered how a fruit fly protein binds and regulates two different types of RNA target sequence. The study, based in part upon research carried out at the U.S. Department of Energy’s Advanced Photon Source (APS), was published in the journal Cell Reports. It may help explain how various RNA-binding proteins, many of which are implicated in cancer and neurodegenerative disease, perform so many different functions in the cell.
There are hundreds of RNA-binding proteins in the human genome that together regulate the processing, turnover, and localization of the many thousands of RNA molecules expressed in cells. These proteins also control the translation of RNA into proteins. RNA-binding proteins are crucial for maintaining normal cellular function, and defects in this family of proteins can lead to disease. For example, RNA-binding proteins are overexpressed in many human cancers, and mutations in some of these proteins have been linked to neurological and neurodegenerative disorders such as amyotrophic lateral sclerosis. "Understanding the fundamental properties of this class of proteins is very relevant," said study co-author Elizabeth Gavis, Professor in the Life Sciences and a professor of molecular biology at Princeton.
Gavis and colleagues are particularly interested in a protein called Glorund (Glo), a type of RNA-binding protein that performs several functions in fruit fly development. This protein was originally identified due to its ability to repress the translation of an RNA molecule called nanos to protein in fly eggs. By binding to a stem structure formed by uracil and adenine nucleotides in the nanos RNA, Glo prevents the production of Nanos protein at the front of the embryo, a step that enables the fly's head to form properly.
Like many other RNA-binding proteins, however, Glo is multifunctional. It regulates several other steps in fly development, apparently by binding to RNAs other than nanos. The mammalian counterparts of Glo, known as heterogeneous nuclear ribonucleoprotein (hnRNP) F/H proteins, bind to RNAs containing stretches of guanine nucleotides known as G-tracts, and, rather than repressing translation, mammalian hnRNP F/H proteins regulate processes such as RNA splicing, in which RNAs are rearranged to produce alternative versions of the proteins they encode.
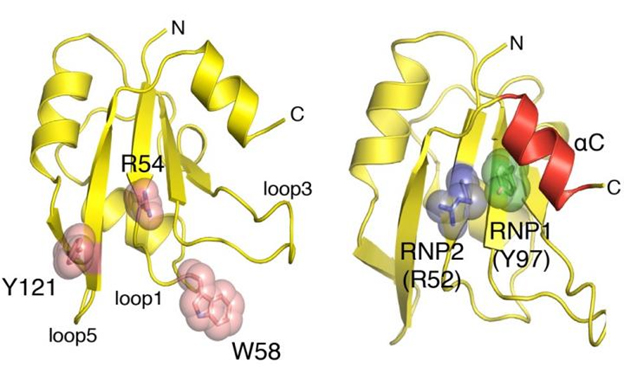
To understand how Glo might bind to diverse RNAs and regulate them in different ways, Gavis and Princeton graduate student Joel Tamayo collaborated with Senior Investigator Traci Tanaka Hall and Research Fellow Takamasa Teramoto from the National Institute of Environmental Health Sciences to determine the crystal structures of Glo's three RNA-binding domains. One of these structures was determined at 1.55-angstrom resolution using high-brightness X-rays provided by the Southeast Regional Collaborative Access Team’s 22-ID-D beamline at the Argonne National Laboratory APS (the APS is an Office of Science user facility). As might be expected, the three domains were highly similar to the corresponding domains of mammalian hnRNP F/H proteins. They retained, for example, the amino acid residues that bind to G-tract RNA, and the researchers confirmed that, like their mammalian counterparts, each RNA-binding domain of Glo can bind to this type of RNA sequence.
However, the researchers also saw something new. "When we looked at the structures, we realized that there were also some basic amino acids that projected from a different part of the RNA-binding domains that could be involved in contacting RNA," Gavis explained. “These residues were within special RNP2 motifs that bind to RNA in related RNA-binding domains,” Hall added. “It seemed reasonable that they might be important for RNA recognition by Glo.”
The researchers found that these basic amino acids mediate binding to uracil-adenine (U-A) stem structures like the one found in nanos RNA. Each of Glo's RNA-binding domains therefore contains two distinct binding surfaces that interact with different types of RNA target sequence. "While there have been examples previously of RNA-binding proteins that carry more than one binding domain, each with a different specificity, this represents the first example of a single domain harboring two different specificities," said Howard Lipshitz, a professor of molecular genetics at the University of Toronto who was not involved in the study.
To investigate which of Glo's two RNA-binding modes was required for its different functions in flies, Gavis and colleagues generated insects carrying mutant versions of the RNA-binding protein. Glo's ability to repress nanos translation during egg development required both of the protein's RNA-binding modes. The researchers discovered that, as well as binding the U-A stem in the nanos RNA, Glo also recognized a nearby G-tract sequence. But Glo's ability to regulate other RNAs at different developmental stages only depended on the protein's capacity to bind G-tracts.
"We think that the binding mode may correlate with Glo's activity towards a particular RNA," said Gavis. "If it binds to a G-tract, Glo might promote RNA splicing. If it simultaneously binds to both a G-tract and a U-A stem, Glo acts as a translational repressor."
The RNA-binding domains of mammalian hnRNP F/H proteins probably have a similar ability to bind two different types of RNA, allowing them to regulate diverse target RNAs within the cell. "This paper represents an exciting advance in a field that has become increasingly important with the discovery that defects in RNA-binding proteins contribute to human diseases such as metabolic disorders, cancer and neurodegeneration," Lipshitz said. "Since these proteins are evolutionarily conserved from fruit flies to humans, experiments of this type tell us a lot about how their human versions normally work or can go wrong."
See: Joel V. Tamayo1, Takamasa Teramoto2, Seema Chatterjee1, Traci M. Tanaka Hall2*, and Elizabeth R. Gavis1**, “The Drosophila hnRNP F/H Homolog Glorund Uses Two Distinct RNA-Binding Modes to Diversify Target Recognition,” Cell Rep. 19, 150 (April 4, 2017).
DOI: 10.1016/j.celrep.2017.03.022
Author affiliations: 1Princeton University, 2National Institutes of Health
Correspondence: **hall4@niehs.nih.gov, *gavis@princeton.edu
The authors would like to thank L. Pedersen and the staff of the Southeast Regional Collaborative Access Team beamlines for assistance with x-ray data collection. This research was supported in part by a National Science Foundation Graduate Research Fellowship awarded to J.V.T. (DGE 1148900), a Japan Society for the Promotion of Science Research Fellowship for Japanese Biomedical and Behavioral Researchers at NIH fellowship awarded to T.T., and grants from the National Institutes of Health to E.R.G. (R01 GM061107), and the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences (T.M.T.H., 1ZIAES050165). This research used resources of the Advanced Photon Source, a U.S. DOE Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02- 06CH11357.
Argonne National Laboratory seeks solutions to pressing national problems in science and technology. The nation's first national laboratory, Argonne conducts leading-edge basic and applied scientific research in virtually every scientific discipline. Argonne researchers work closely with researchers from hundreds of companies, universities, and federal, state and municipal agencies to help them solve their specific problems, advance America's scientific leadership and prepare the nation for a better future. With employees from more than 60 nations, Argonne is managed by UChicago Argonne, LLC for the U.S. Department of Energy's Office of Science.
The U.S. Department of Energy's Office of Science is the single largest supporter of basic research in the physical sciences in the United States and is working to address some of the most pressing challenges of our time. For more information, visit the Office of Science website.