The original University of Wisconsin-Madison press release by Eric Hamilton can be read here.
As you bite into your next peanut butter and jelly sandwich, chew on this: The peanut you’re eating has a secret. It’s a subtle one. The peanut and its kin — legumes — have not one, but two ways to make the amino acid tyrosine, one of the 20 required to make all of its proteins, and an essential human nutrient. That might seem small, but why this plant family has a unique way to make such an important chemical building block is a mystery that extends back to the 1960s and is one that has captured the attention of Hiroshi Maeda, a professor of botany at the University of Wisconsin–Madison (UWM). In new research abetted by high-brightness x-ray beams from the U.S. Department of Energy’s (DOE’s) Advanced Photon Source (APS) Maeda and his collaborators report in Nature Chemical Biology how the legume family evolved its second tyrosine pathway.
“We’re interested in plant chemistry, trying to understand how plants make so many different chemical compounds, many of which are important to our human society as food, fiber, feed, fuel, medicine — so many things,” says Maeda.
Those important molecules start from simpler compounds, like tyrosine, which is the precursor of morphine and countless other interesting and useful chemicals.
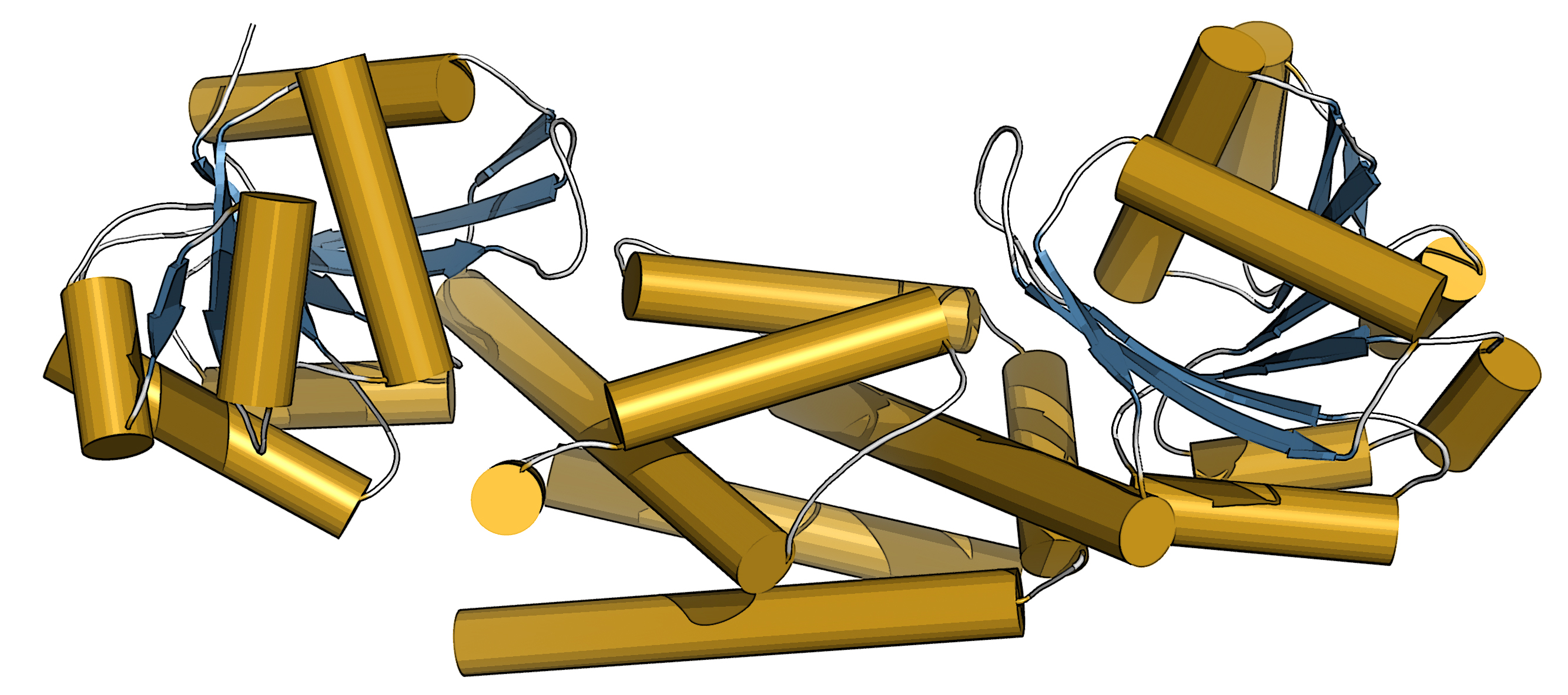
Employing x-ray diffraction data collected at the Structural Biology Center Collaborative Access Team (SBC-CAT) beamline 19-ID-D at the APS, the team of researchers from UWM and Washington University in St. Louis determined the structure of a new plant enzyme, one which could be a useful tool to biotechnologists trying to control the production of tyrosine and its derivatives. And they tied a major evolutionary change in plant metabolism to a single mutation in the new enzyme. (The APS is an Office of Science user facility at Argonne National Laboratory.)
In the 1960s and ’70s, scientists surveyed the plant world to find how they made key metabolic compounds, like amino acids. While all plants used one pathway, known as ADH, to make tyrosine, the legume family — peas, beans, peanuts — uniquely added a second, called PDH, which was otherwise found only in microbes. Nobody knew why, and the problem was set aside.
But two years ago, Maeda and his graduate student, Craig Schenck, dusted off the old mystery. Digging in, they discovered the genes responsible for making tyrosine. They found that the legumes had evolved their PDH enzymes from existing ADH ones, just before peanuts and peas evolved into separate lineages. The sister enzymes were very similar, which meant only a small number of changes could account for how the ADH enzymes evolved into the PDH ones. But there were still too many changes to test one by one to see which ones had an effect.
Then Maeda received a call from Joe Jez, a biochemist at Washington University in St. Louis. With Jez’s student Cynthia Holland, the two teams collaborated to purify the PDH enzyme of soybean, a legume, and determine its three-dimensional structure. With the structure of PDH in hand, Schenck could see that over evolutionary time, only a couple mutations had occurred at the site where the chemical reactions take place. Instead of dozens of mutations to try, he had only two.
Schenck found that by changing a single amino acid in the center of the enzyme, he was able to largely convert the soybean PDH enzyme back into its ancestor ADH enzyme. The switch worked for enzymes from multiple species, and worked in reverse: Schenck could give ADH enzymes from non-legume plants PDH-like characteristics.
Maeda and Schenck had discovered that legumes evolved a novel way to make an important chemical mostly by stumbling on a single, crucial switch.
“The most surprising result is that a single residue really played a major role in switching to make this legume-specific enzyme,” says Maeda. “And that raises an interesting question of why other groups of plants never evolved this unique enzyme. Because just with random chance, perhaps this mutation occurred but was never maintained.”
Just why legumes held on to their new tyrosine pathway, and what advantage it might provide, will require more work.
Another takeaway, says Maeda, is that the same switch that turns ADH enzymes into PDH ones shuts off the ability of tyrosine to inhibit the function of the enzyme. Although this kind of self-regulation is normally useful for cells, Maeda thinks that the PDH insensitivity to tyrosine could be a boon for helping to produce more tyrosine, and its useful derivatives, in systems like yeast or engineered plants.
“The thought is that opium poppy, for example, is making tyrosine through a standard ADH pathway that is likely inhibited by tyrosine,” explains Schenck, discussing possible applications for the new research. “If we can introduce an enzyme that isn’t inhibited by tyrosine, maybe we can increase the total pool of the precursor tyrosine for increasing morphine production. It may be a useful tool going forward in other plant species or even in microbes.”
See: Craig A. Schenck1, Cynthia K. Holland2, Matthew R. Schneider1, Yusen Men1, Soon Goo Lee2, Joseph M. Jez2, and Hiroshi A. Maeda1*, “Molecular basis of the evolution of alternative tyrosine biosynthetic routes in plants,” Nat. Chem. Biol. 13, 1029 (13 September 2017). DOI: 10.1038/nchembio.2414
Author affiliations: 1University of Wisconsin-Madison, 2Washington University in St. Louis
Correspondence: *maeda2@wisc.edu
This work was supported by the National Science Foundation (NSF) IOS-1354971 to H.A.M. and MCB-1614539 to J.M.J. C.K.H. was supported by the NSF Graduate Research Fellowship Program (DGE-1143954). SBC-CAT is operated by UChicago Argonne, LLC, for the U.S. DOE Office of Science Biological and Environmental Research Program under Contract No. DE-AC02-06CH11357. This research used resources of the Advanced Photon Source, a U.S. DOE Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
Argonne National Laboratory seeks solutions to pressing national problems in science and technology. The nation's first national laboratory, Argonne conducts leading-edge basic and applied scientific research in virtually every scientific discipline. Argonne researchers work closely with researchers from hundreds of companies, universities, and federal, state and municipal agencies to help them solve their specific problems, advance America's scientific leadership and prepare the nation for a better future. With employees from more than 60 nations, Argonne is managed by UChicago Argonne, LLC for the U.S. Department of Energy's Office of Science.
The U.S. Department of Energy's Office of Science is the single largest supporter of basic research in the physical sciences in the United States and is working to address some of the most pressing challenges of our time. For more information, visit the Office of Science website.