The original John Hopkins University School of Medicine press release can be read here.
Researchers at Johns Hopkins University School of Medicine report they have laid the foundation to develop novel antibiotics that work against incurable, antibiotic-resistant bacteria like tuberculosis by targeting an enzyme essential to the production and integrity of bacterial cell walls. The findings, aided by research at the U.S. Department of Energy’s Advanced Photon Source (APS), suggest that antibiotic drugs specifically targeting the recently discovered LD-transpeptidase enzyme, which is needed to build bacterial cell walls in some bacteria, could potentially cure many antibiotic-resistant infections.
An additional implication of the research, the team says, is that drugs targeting the enzyme could offer quicker, cheaper and more easily accessible treatment against tuberculosis, a disease that still kills more people worldwide than any other infection, according to the U.S. Centers for Disease Control and Prevention.
A summary of the findings was published on November 7, 2016, in Nature Chemical Biology.
"The complexity of TB treatment and growing prevalence of antibiotic resistance is a serious threat to public health," said article lead author Gyanu Lamichhane, associate professor of medicine at the Johns Hopkins University School of Medicine. His team joined with the research team of Craig Townsend, the Alsoph H. Corwin Professor of Chemistry at The Johns Hopkins University's Krieger School of Arts and Sciences, and researchers from Rutgers University, Collaborations in Chemistry, and Argonne National Laboratory to tackle this complex issue. "Our research offers steps toward the design of new antibiotics that attack a previously untargeted bacterial enzyme," said co-author Townsend.
At the root of their investigation, Lamichhane said, is the fact than more than half of antibiotics prescribed today are of a class called beta-lactams, which work by interrupting the function of the DD-transpeptidase enzyme that creates bacterial cell walls. Without it, bacteria quickly die. However, in 2005, a team of researchers found a second wall-building enzyme, LD-transpeptidase, that allows bacteria like the ones that cause TB to survive antibiotic treatments. "We looked at the structure of LD-transpeptidase, thought about how it works and started making new compounds that could be used against it," said Townsend.
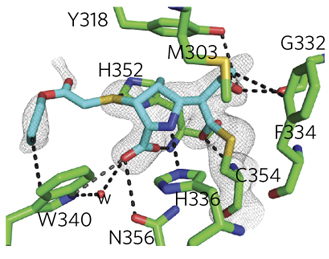
Pankaj Kumar, co-author and postdoctoral fellow in infectious diseases at the Johns Hopkins University School of Medicine, began the research in the new study by extracting LD-transpeptidase from many species of bacteria and examining its detailed molecular structure with a sophisticated imaging system known as protein x-ray crystallography using the Structural Biology Center 19-ID x-ray beamline at the Argonne APS (which is an Office of Science user facility) in collaboration with Dr. Stephan Ginell whose contribution was instrumental in generating the structure data reported in this work.
By analyzing the enzyme's structure, Johns Hopkins researchers were able to design new compounds in the carbapenem group, a subclass of the beta-lactam antibiotics that bind to the LD-transpeptidase wall-building enzyme and stop its function.
In live bacterial cultures, the carbapenems were shown by Lamichhane's and Townsend's groups to stop the enzyme's wall-building activity. The new compounds were even effective against the ESKAPE pathogens, a group of six bacterial species that the Centers for Disease Control and Prevention has identified as a threat because of their propensity for developing antibiotic resistance.
Following these successes, co-author Amit Kaushik, a postdoctoral fellow in infectious diseases at the Johns Hopkins University School of Medicine, tested two carbapenems in vivo against TB in mice infected with TB.
Researchers infected mice with tuberculosis bacteria and separated them into different treatment groups. The rodents' lungs were sampled periodically over a period of three weeks, and the results showed that even without use of classic TB antibiotic treatments, the new carbapenems, specifically biapenem, cured TB infection in mice.
"Our data show that the carbapenems successfully treated TB infections alone by attacking the right enzyme," said Lamichhane.
Townsend and Lamichhane say the focus of their research is now on creating variations of their original compound that are designed to target specific bacteria. Many commonly prescribed antibiotics today work on a broad spectrum of bacterial species, meaning that in addition to killing off bad bacteria, they also destroy the friendly bacteria our bodies need to function normally and whose destruction can cause dangerous side effects. Lamichhane believes that the future of antibiotic treatments relies on our ability to be good antimicrobial stewards and treat specific bacteria without affecting our bodies' natural microbiome. Not only will this cut down on antibiotic side effects, but it will also slow the development of antibiotic resistance in the bacterial species not being targeted.
The researchers are now in the process of initiating clinical trials to test the safety and efficacy of some of these new compounds.
Antibiotic resistance has been an ever-present threat since the discovery of penicillin in 1928. Scientists and physicians have historically kept up with resistant bacteria through the frequent introduction of new antibiotic treatments. However, the research and development of antibiotics plummeted in the 1980s as other drugs became more profitable to produce. Because of the decreased incentive to invest in new antibiotics and the liberal use of antibiotics we already have, many bacterial species have quickly outpaced our ability to treat them. The Centers for Disease Control and Prevention estimates that annually, 2 million illnesses and 23,000 deaths are caused by antibiotic-resistant bacteria in the United States alone, and that 70% of hospital-acquired infections are now antibiotic-resistant.
See: Pankaj Kumar1, Amit Kaushik1, Evan P, Lloyd1, Shao-Gang Li2, Rohini Mattoo1, Nicole C. Ammerman1, Drew T. Bell1, Alexander L Perryman2, Trevor A. Zandi1, Sean Ekins3, Stephan L. Ginell4, Craig A. Townsend1, Joel S. Freundlich2, and Gyanu Lamichhane1*, “Non-classical transpeptidases yield insight into new antibacterials,” Nat. Chem. Biol., advance online publication, 07 November 2016. DOI: 10.1038/nchembio.2237
Author affiliations: 1Johns Hopkins University, 2Rutgers University Medical School, 3Collaborations in Chemistry, 4Argonne National Laboratory
Correspondence: *lamichhane@jhu.edu
This study was supported by National Institutes of Health awards R21AI111739 and DP2OD008459 to G.L. The Structural Biology Center at the Advanced Photon Source, Argonne, is operated by UChicago Argonne, LLC, for the U.S. Department of Energy (DOE) Office of Biological and Environmental Research under contract DE-AC02-06CH11357. This research used resources of the Advanced Photon Source, a U.S. DOE Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
Argonne National Laboratory seeks solutions to pressing national problems in science and technology. The nation's first national laboratory, Argonne conducts leading-edge basic and applied scientific research in virtually every scientific discipline. Argonne researchers work closely with researchers from hundreds of companies, universities, and federal, state and municipal agencies to help them solve their specific problems, advance America's scientific leadership and prepare the nation for a better future. With employees from more than 60 nations, Argonne is managed by UChicago Argonne, LLC for the U.S. Department of Energy's Office of Science.
The U.S. Department of Energy's Office of Science is the single largest supporter of basic research in the physical sciences in the United States and is working to address some of the most pressing challenges of our time. For more information, visit the Office of Science website.