With an assist from research at the U.S. Department of Energy’s Advanced Photon Source (APS), an Office of Science user facility at Argonne, Caltech scientists have produced the most detailed map yet of the massive protein machine that controls access to the DNA-containing heart of the cell.
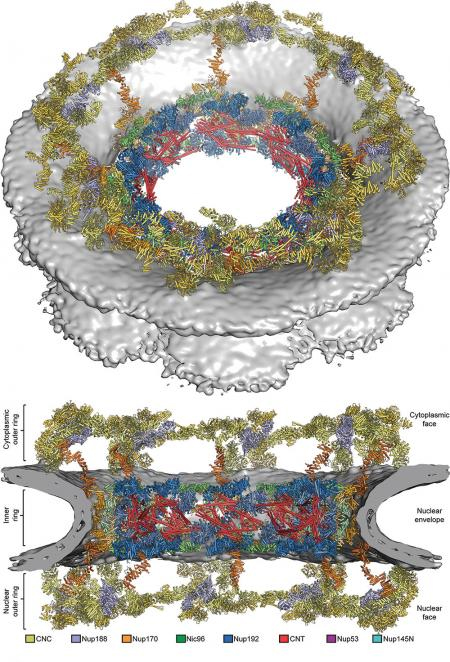
In a new study, the team led by André Hoelz, an assistant professor of biochemistry at Caltech, reports the successful mapping of the structure of the symmetric core of the nuclear pore complex (NPC), a cellular gatekeeper that determines what molecules can enter and exit the nucleus, where a cell's genetic information is stored.
The study appeared in the journal Science, and was featured on the cover.
The findings are the culmination of more than a decade of work by Hoelz's research group and could lead to new classes of medicine against viruses that subvert the NPC in order to hijack infected cells and that could treat various diseases associated with NPC dysfunction.
"The methods that we have been developing for the last 12 years open the door for tackling other large and flexible structures like this," said Hoelz. "The cell is full of such machineries but they have resisted structural characterization at the atomic level…"
…As described in the Science paper, Hoelz's research group now has solved the crystal structures of the symmetric core's inner ring and determined where all of the rings' pieces fit in the NPC's overall structure.
To do this, the team had to first generate a complete "biochemical interaction map" of the entire symmetric core. Akin to a blueprint, this map describes the interconnections and interactions of all of the proteins, as part of a larger cellular machine. The process involved genetically modifying bacteria to produce purified samples of each of the 19 different protein subunits of the NPC's symmetric core and then combining the fragments two at a time inside a test tube to see which adhered to each other.
The team then used the completed interaction map as a guide for identifying the inner ring's key proteins and employed x-ray crystallography to determine the size, shape, and position of all of their atoms. The team analyzed thousands of samples at Caltech's Molecular Observatory — a facility developed with support from the Gordon and Betty Moore Foundation that includes an x-ray beam line at the Stanford Synchrotron Radiation Laboratory — and the National Institute of General Medical Sciences and National Cancer Institute/X-ray Science Division x-ray beamline at the APS.
"We now had a clear picture of what the key jigsaw pieces of the NPC looked like and how they fit together," said Daniel Lin, a graduate student in Hoelz's lab and one of two first authors on the study.
See: Daniel H. Lin, Tobias Stuwe, Sandra Schilbach, Emily J. Rundlet, Thibaud Perriches, George Mobbs, Yanbin Fan, Karsten Thierbach, Ferdinand M. Huber, Leslie N. Collins, Andrew M. Davenport, Young E. Jeon, and André Hoelz*, “Architecture of the symmetric core of the nuclear pore,” Science 352(6283) (15 APRIL 2016). DOI: 10.1126/science.aaf1015
Author affiliation: California Institute of Technology
Correspondence: *hoelz@caltech.edu
D.H.L and A.M.D. were supported by a National Institutes of Health (NIH) Research Service Award (5 T32 GM07616). D.H.L. was also supported by an Amgen Graduate Fellowship through the Caltech-Amgen Research Collaboration. T.S. was supported by a postdoctoral fellowship from the Deutsche Forschungsgemeinschaft. F.M.H. was supported by a doctoral fellowship from the Boehringer Ingelheim Fonds. Y.F. was supported by a visiting doctoral student scholarship from the China Scholarship Council. A.H. was supported by Caltech startup funds, the Albert Wyrick V Scholar Award from the V Foundation for Cancer Research, the 54th Mallinckrodt Scholar Award from the Edward Mallinckrodt Jr. Foundation, a Kimmel Scholar Award from the Sidney Kimmel Foundation for Cancer Research, a Camille-Dreyfus Teacher Scholar Award, and NIH grant R01-GM111461, and he is an inaugural Heritage Principal Investigator of the Heritage Medical Research Institute. GM/CA has been funded in whole or in part with federal funds from the National Cancer Institute (grant ACB-12002) and the National Institute of General Medical Sciences (grant AGM-12006). Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy (DOE) Office of Science-Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393). This research used resources of the Advanced Photon Source, a U.S. DOE Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
Argonne National Laboratory seeks solutions to pressing national problems in science and technology. The nation’s first national laboratory, Argonne conducts leading-edge basic and applied scientific research in virtually every scientific discipline. Argonne researchers work closely with researchers from hundreds of companies, universities, and federal, state and municipal agencies to help them solve their specific problems, advance America’s scientific leadership and prepare the nation for a better future. With employees from more than 60 nations, Argonne is managed by UChicago Argonne, LLC for the U.S. Department of Energy’s Office of Science.
The U.S. Department of Energy’s Office of Science is the single largest supporter of basic research in the physical sciences in the United States and is working to address some of the most pressing challenges of our time. For more information, visit the Office of Science website.