Animals can rapidly adapt to changes in the environment through subtle changes to the structure of their bodies’ proteins. This process is referred to as “post-translational modification,” which can impact the function, structure, stability, and location of proteins involved in essential cellular processes, including translation, transcription and metabolism. Such modifications give organisms the flexibility to adapt rapidly to changes in their environment. One ubiquitous form of post-translational modifications is acetylation, the donation of an acetyl functional group from acetyl coenzyme A to a lysine residue. X-ray crystallography studies at the U.S. Department of Energy's Advanced Photon Source (APS), an Office of Science user facility, helped reveal that acetylation of Escherichia coli proteins can also be non-enzymatic, specific, and occurs at both active sites and co-factor binding sites. Additional experiments demonstrated that acetylation is likely capable of modifying the function of important enzymes, a finding that could have implications for the engineering of bacteria for specific applications in a variety of fields such as energy production, medicine, and environmental remediation.
Acetylation can have a variety of impacts on cellular processes, including altering protein size, DNA binding affinity, and enzymatic activity. While acetylation has been well documented in eukaryotes, only in the past few years have researchers obtained genetic, mass spectrometric, chemical, and structural evidence of acetylation of thousands of lysine residues in hundreds of E. coli proteins, suggesting that acetylation is every bit as critical and widespread in bacteria as it is in eukaryotes. Multiple threads of evidence also indicated that the molecule acetyl phosphate can function as an acetyl donor in bacteria, along with the better known acetyl coenzyme A.
One type of post-translational modification is Ne-lysine acetylation, in which an enzyme catalyzes the donation of an acetyl group from acetyl-coenzyme A (acCoA) to the ε-amino group of a lysine residue within a protein. This is known to be very common in eukaryotes, but has only recently been shown to occur in bacteria. However, the prevalence of acetylation and its mechanism(s) had not yet been examined in detail.
The research team in this study, which included scientists from Northwestern University Feinberg School of Medicine; Loyola University; the Buck Institute for Research on Aging; Northwestern University; and the University of California, San Francisco had previously discovered an E. coli mutant with markedly different acetylation patterns compared to wild type. The mutant accumulates a molecule called acetyl phosphate (acP), leading the scientists to hypothesize that acP regulates acetylation. In the current study, increased acetylation in the presence of glucose was observed in the E. coli acP-accumulating mutants. In contrast, acetylation decreased, but was not eliminated, in other mutants that could not synthesize acP, results that confirmed the team’s hypothesis.
Mass spectrometry and Western immunoblot analysis revealed thousands of acetylation sites on hundreds of lysine proteins across a handful of E. coli strains. Most of the acetylated proteins contained one or two acetylated lysines, but some contained as many as 24. These experiments also verified the existence of both acP-dependent and acP-independent acetylation. Other mass spectrometry experiments indicated that acP can act directly as an acetyl donor without the need of an enzyme such as lysine acetyltransferase. Because acP is a small molecule it can access active sites that most enzymes cannot, leading the researchers to speculate that acP could have a role in acetylating, and thereby inhibiting, molecules such as RNA polymerase.
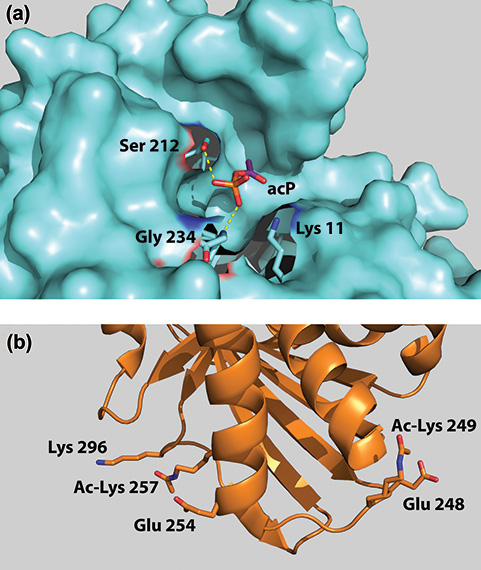
Searching through the gene function database PANTHER revealed that the most common functions of the observed acetylated lysines were metabolic in nature. AcP-driven acetylation was also found to target proteins involved in translation and transcription. Using the Life Sciences Collaborative Access Team (LS-CAT) 21-ID-D, 21-ID-F, and 21-ID-G beamlines at the Argonne APS, the researchers derived x-ray crystal structures of two of these proteins, triosephosphate isomerase (TpiA) and glyceraldehyde-3-phosphate dehydrogenase (GapA), both of which are involved in glycolysis [Fig.1(a) and (b)]. Comparing the three-dimensional structures before and after briefly bathing the proteins in acP confirmed that acP can bind lysine in the absence of an enzyme and can do so at both active sites and co-factor binding sites.
Given the geometry of acP binding sites observed in the crystal structures, as well as previously reported effects of acetylation in other organisms, the research team inferred that acetylation would likely inhibit the enzymatic activity of both TpiA and GapA.
The finding that acetylation of bacterial proteins involved in crucial cellular processes is commonplace, and likely carries functional consequences, opens avenues for custom engineering microbes used in a variety of fields. The research team plans to investigate how acetylation impacts protein function, as well as determine how the process is regulated within living bacteria. — Chris Palmer
See: Misty L. Kuhn1, Bozena Zemaitaitis2, Linda I. Hu2, Alexandria Sahu3, Dylan Sorensen3, George Minasov1, Bruno P. Lima2‡, Michael Scholle4, Milan Mrksich4, Wayne F. Anderson1, Bradford W. Gibson3,5, Birgit Schilling3, and Alan J. Wolfe2*, “Structural, Kinetic and Proteomic Characterization of Acetyl Phosphate-Dependent Bacterial Protein Acetylation,” PLoS One 9(4), e94816 (2014). DOI: 10.1371/journal.pone.0094816
Author affiliations: 1Northwestern University Feinberg School of Medicine, 2Loyola University Chicago, 3Buck Institute for Research on Aging, 4Northwestern University, 5University of California, San Francisco‡Present address: University of California, Los Angeles
Correspondence: *awolfe@luc.edu
This work was supported by grants from the National Institutes of Health (NIH), National Institute of Allergy and Infectious Diseases (NIAID), and National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK); including R24 DK085610 (NIDDK, to B.W.G.), R01 GM066130 (National Institute of General Medical Sciences, to A.J.W.), Department of Health and Human Services Contracts HHSN272200700058C and HHSN272201200026C (NIAID, NIH, Department of Health and Human Services to W.F.A.) and U54CA151880 (National Cancer Institute, to M.M.). The use of LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and Michigan Technology Tri-Corridor (Grant085P100817). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
Argonne National Laboratory is supported by the Office of Science of the U.S. Department of Energy. The Office of Science is the single largest supporter of basic research in the physical sciences in the United States, and is working to address some of the most pressing challenges of our time. For more information, please visit science.energy.gov.