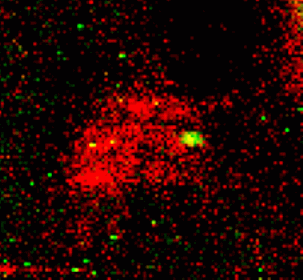
Metals such as copper, zinc, and iron are important nutrients to all life. The special properties of these elements that make them so useful in technologies including batteries and catalysts – for example, having multiple stable oxidations states under ambient conditions – also make them useful to living organisms.
With over a third of all proteins thought to bind metals, knowing which metals are bound and how that binding changes in response to the environment could have big implications.
For instance, the biological mismanagement of metals is involved in many diseases, including Lou Gehrig’s disease, Wilson and Menkes disease, and possibly even Alzheimer’s disease.
Metals are also an environmental toxin, such as the hexavalent chromium featured in the movie Erin Brockovich, and they are used in drugs, like the platinum in cisplatin that treats prostate cancer.
Developing an approach for making determinations about the relationship of metals and proteins is complex because the experimental methods that are routinely used to identify proteins, for example denaturing gel electrophoresis, can also remove metals that might be bound to them.
Scientists working at the Department of Energy Office of Science’s Advanced Photon Source (APS) at Argonne have made great strides in imaging metals within cells. Using the x-ray imaging capabilities afforded by the APS, researchers have seen, often for the first time, where the metals reside inside cells and tissues. These capabilities have allowed researchers to see how the elemental content of bacteria change upon adhesion, fluxes of zinc in egg cells upon fertilization, and changes in the locations where copper is stored in a cell during the growth of blood vessels.
But many of the images that have been acquired led to new questions: Are these metals required for the activity of proteins? Which proteins are binding with which metals inside the cell?
Now, a team of researchers from the Worcester Polytechnic Institute and Argonne carrying out research at the APS have developed a new experimental approach that not only detects and distinguishes metals in proteins, but also characterizes the proteins that bind the metals, without removing them. This work, which was featured on the cover of the journal Metallomics, utilized x-ray fluorescence imaging (XRF) at X-ray Science Division (XSD) beamline 8-BM-B of the APS.
Employing modified native two-dimensional gel electrophoresis, the researchers were able to separate proteins from the organisms S. oneidensis, a bacterium that can reduce poisonous heavy metal and can live in both environments with or without oxygen, and P. aeruginosa, a common bacterium that can cause disease in animals, including humans, and that is found in soil, water, skin flora, and most man-made environments throughout the world. Then, using XRF, the team quantitatively measured the amount of sulfur, iron, and zinc at every point of the two-dimensional (2-D) separation, pinpointed the location of proteins that had metals bound to them, and determined the identity of these proteins utilizing mass spectrometry.
The approach enabled the research team to identify a novel protein (PA5217) as a zinc-binding protein in P. aeruginosa.
Their finding highlights how this method not only determines changes in metal occupancy, but also identifies the associated protein.
Now that this new technique is developed, questions raised by images of the metals in cells can be studied further.
Native 2-D gel electrophoresis separation is accessible to most laboratories, and resources for 2-D XRF imaging are available at the APS.
This development will help researchers begin to identify which of the one-third of proteins that are thought to bind metals actually do, and what roles they play in life.
See: Daniel Raimunda1, Tripti Khare2, Carol Giometti2, Stefan Vogt2, José M. Argüello1, and Lydia Finney2*, “Identifying metalloproteins through X-ray fluorescence mapping and mass spectrometry,” Metallomics 4, 921 (2012). DOI:10.1039/c2mt20095c
Author affiliations: 1Worcester Polytechnic Institute, 2Argonne National Laboratory
Correspondence: *lfinney@aps.anl.gov
This work was supported by National Science Foundation grant MCB-0743901 (JMA), and U.S. Department of Agriculture-National Institute of Food and Agriculture grant 2010-65108-20606 (JMA). Use of the Advanced Photon Source at Argonne National Laboratory was supported by the U.S. Department of Energy’s Office of Science under Contract No. DE-AC02-06CH11357.
The Advanced Photon Source at Argonne National Laboratory is one of five national synchrotron radiation light sources supported by the U.S. Department of Energy’s Office of Science to carry out applied and basic research to understand, predict, and ultimately control matter and energy at the electronic, atomic, and molecular levels, provide the foundations for new energy technologies, and support DOE missions in energy, environment, and national security. To learn more about the Office of Science x-ray user facilities, visit http://science.energy.gov/user-facilities/basic-energy-sciences/.
Argonne National Laboratory seeks solutions to pressing national problems in science and technology. The nation's first national laboratory, Argonne conducts leading-edge basic and applied scientific research in virtually every scientific discipline. Argonne researchers work closely with researchers from hundreds of companies, universities, and federal, state and municipal agencies to help them solve their specific problems, advance America's scientific leadership and prepare the nation for a better future. With employees from more than 60 nations, Argonne is managed by UChicago Argonne, LLC for the U.S. Department of Energy's Office of Science.