The structure of a key protein from the virus that caused last year's "swine flu" influenza epidemic has been solved by researchers using two U.S. Department of Energy synchrotron light sources, including the Advanced Photon Source at Argonne. The structure reveals that the virus shares many features with influenza viruses common in the early 20th century, helping to explain why, in general, older individuals have been less severely affected by the recent outbreak than younger ones. The information should be useful for scientists and public health officials as they respond to current and future pandemics.
In the study, first published in the March 25, 2010, issue of Science Express, the researchers from The Scripps Research Institute, Vanderbilt University, and the Mount Sinai School of Medicine describe the structure of the hemagglutinin (the influenza virus envelope protein) from the H1N1 swine flu virus that triggered the pandemic in 2009 and is still circulating in the human population. The team then compared the swine flu hemagglutinin protein with a range of different human H1N1 flu viruses in the past century.
“Parts of the 2009 virus are remarkably similar to human H1N1 viruses circulating in the early 20th century,” said Scripps Research Professor Ian Wilson, who was the senior author of the study. “Our findings provide strong evidence that exposure to earlier viruses has helped to provide some people with immunity to the recent influenza pandemic.”
Influenza is a common viral infection of the lungs that affects millions of people annually and is a leading cause of death in the United States, contributing to around 50,000 deaths per year. Serious influenza outbreaks such as the deadly "Spanish flu" of 1918 have occurred when a virus adapted to birds jumps directly into humans or reasserts and infects another species, such as the pig, and then jumps into humans. Similar outbreaks occurred in 1957 and 1968.
"For a pandemic to occur, there needs to be a native population, whose immune systems have not learned to recognize the virus and who can be infected," explained Rui Xu, a research associate in the Scripps Research Wilson lab who was first author of the paper with graduate student Damian Ekiert, also of the Wilson lab. "A pandemic outbreak is different from the seasonal flu, in which existing flu viruses circulate in the human population, gradually mutating as time goes on."
The most recent influenza outbreak, dubbed the "swine flu" by the media due to its recent origin in pigs, was first reported in Mexico in April 2009. The virus has now spread worldwide, and has contributed to at least 16,000 deaths, according to the World Health Organization. A vaccine is now available, but the virus remains a public health concern.
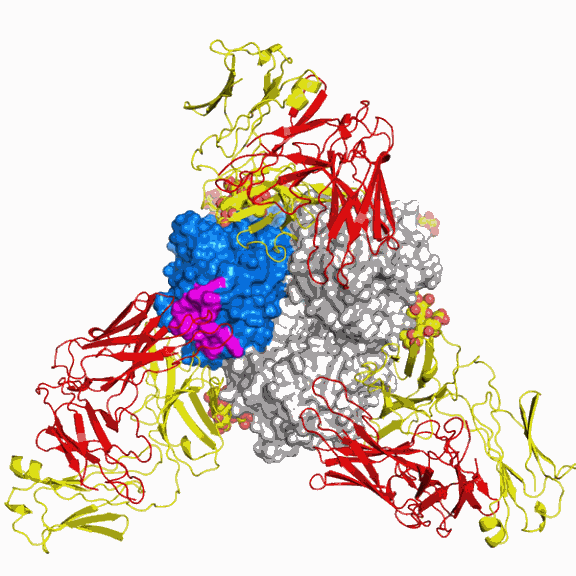
Collaborating with colleagues at Mount Sinai School of Medicine, who provided a clone of the major surface antigen from the emerging virus, A/California/04/2009 (CA04), the researchers utilized x-ray crystallography at the GM/CA-CAT 23-ID-B beamline at the Advanced Photon Source and beamline 9-2 at the Stanford Synchrotron Radiation Lightsource (SLAC National Accelerator Laboratory) for the structure determinations. The scientists chose to focus on the structure of the virus's hemagglutinin, a protein that is abundantly displayed on the viral surface. In addition to enabling the virus to infect cells of the host organism, hemagglutinin is the main antigenic determinant on the virus. In other words, it is what the immune system primarily recognizes and responds to by making antibodies (a type of immune molecule) and mounting an immune defense. Vulnerability to an individual influenza infection depends on how well a person's immune system recognizes the hemagglutinin.
The scientists' initial experiments went extraordinarily well, and by June, the team was able to reconstruct the structure of the swine flu hemagglutinin. But what did the structure mean?
That's when the hard work began.
"One of the interesting aspects of the study to us was that the H1N1 subtype was already circulating in humans," said Xu. "That is the first time that we have seen such a phenomenon. How could the same sub-type of influenza virus induce a new pandemic?"
Comparing the 2009 hemagglutinin protein with the hemagglutinin of other influenza samples, including the 1918 flu (a structure that Wilson and colleagues solved six years ago), helped provide answers. For the analysis, the scientists used all known human H1N1 strains between 1918 and 1957, and representative strains since 1977.
The researchers found that while much of the hemagglutinin three-dimensional structure had been maintained among the different viruses, the amino acids (protein building blocks) on the viral surface were substantially different in the 2009 virus from seasonal strains. This could enable the virus to initially evade detection by the immune system.
Strikingly, the scientists also found that one area of the hemagglutinin, called antigenic site Sa, was highly similar between the 2009 and the 1918 viruses. The similarity of the Sa site for the two viruses suggested that some individuals might be able to mount an immune response that could neutralize either virus.
That would have remained an educated guess if Lady Luck hadn't intervened.
In another flu project ongoing in the Wilson lab, Ekiert was working to determine the structure of an antibody that neutralized the 1918 influenza virus. The antibody had been isolated from a survivor of the 1918 Spanish flu. "As more information became available, the 1918 antibody suddenly became relevant to the swine flu study," said Ekiert.
Could the particular antibody that Ekiert was working with, called 2D1, not only be effective against the 1918 virus, but also act against the 2009 swine flu?
A study recently published in the Journal of Virology with researchers at Vanderbilt University, who are collaborators on this present work, showed that, indeed, mice challenged with the 2009 virus are protected by the administration of the antibody against the 1918 virus. The current Science Express study provides the structure of the 2D1 antibody in complex with the 1918 virus and addresses how this protection occurs.
"There is a huge divergence among different influenza viruses," said Ekiert, "so that exposure to one won't confer protection against another. However, this study shows that prior exposure to viruses that were around decades ago can provide some protection against infection against a newly emerging pandemic."
See: Rui Xu1 , Damian C. Ekiert1, Jens C. Krause2, Rong Hai3, James E. Crowe, Jr.2, Ian A. Wilson1*, “Structural Basis of Preexisting Immunity to the 2009 H1N1 Pandemic Influenza Virus, Published Online March 25, 2010. Science DOI: 10.1126/science.1186430
Author affiliations: 1The Scripps Research Institute, 2Vanderbilt University Medical Center, 3Mount Sinai School of Medicine
Correspondence: *wilson@scripps.edu
The original Scripps Research Institute press release can be found here.
This work was supported by the National Institutes of Health (NIH), the Skaggs Institute for Chemical Biology, and predoctoral fellowships from the Achievement Rewards for College Scientists Foundation and the NIH Molecular Evolution Training Program. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357.
Argonne National Laboratory seeks solutions to pressing national problems in science and technology. The nation's first national laboratory, Argonne conducts leading-edge basic and applied scientific research in virtually every scientific discipline. Argonne researchers work closely with researchers from hundreds of companies, universities, and federal, state and municipal agencies to help them solve their specific problems, advance America's scientific leadership and prepare the nation for a better future. With employees from more than 60 nations, Argonne is managed by UChicago Argonne, LLC for the U.S. Department of Energy's Office of Science.