A detailed map that pinpoints the location of every atom in a 450-million-year-old resurrected protein reveals the precise evolutionary steps needed to create the molecule's modern version, according to researchers from the University of North Carolina at Chapel Hill and the University of Oregon, who determined the atomic structure of the ancient protein by utilizing a South East Regional Collaborative Access Team beamline at the Argonne Advanced Photon Source (APS), and the Stanford Synchrotron Radiation Laboratory beamline 11-1.
Until now, scientists trying to unravel the evolution of the proteins and other molecules necessary for life have worked backwards, making educated guesses based on modern human body chemistry. By moving forward from an ancient protein, the team laid out the step-by-step progression required to reach its current form and function.
This research afforded the first opportunity to see the precise mechanisms by which evolution molded a tiny molecular machine at the atomic level, and to reconstruct the order of events by which history unfolded. The study is in a paper that appeared online August 16 in Science Express.
A detailed understanding of how proteins – the workhorses of every cell – have evolved has long eluded evolutionary biologists, in large part because ancient proteins have not been available for direct study. Project leader Joe Thornton, an evolutionary biologist at the University of Oregon, and Jamie Bridgham, a postdoctoral scientist in his lab, used state-of-the-art computational and molecular techniques to re-create the ancient progenitors of an important human protein.
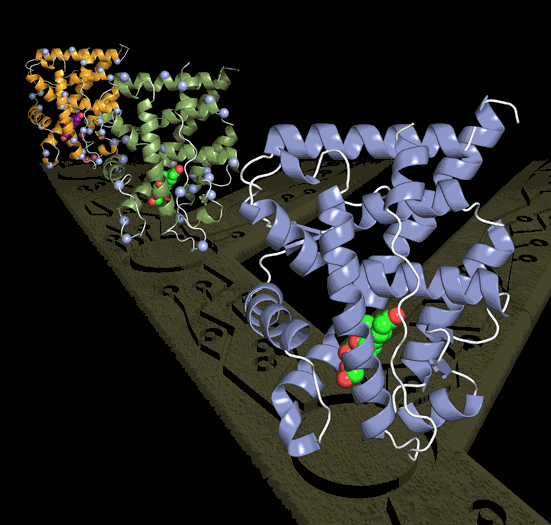
Thornton then collaborated with University of North Carolina biochemists Eric Ortlund, lead author in the study, and Matthew Redinbo, who used ultra-high-energy x-rays from the APS to chart the precise position of each of the 2,000 atoms in the ancient proteins. The groups then worked together to trace how changes in the protein's atomic architecture over millions of years caused it to evolve a crucial new function – uniquely responding to the hormone that regulates stress.
The researchers focused on the glucocorticoid receptor (GR), a protein in humans and other vertebrates that allows cells to respond to the hormone cortisol, which regulates the body's stress response. The scientists' goal was to understand the process of evolution behind the GR's ability to specifically interact with cortisol. They used computational techniques and a large database of modern receptor sequences to determine the ancient GR's gene sequence from a time just before and just after its specific relationship with cortisol evolved. The ancient genes – which existed more than 400 million years ago – were then synthesized, expressed, and their structures determined using x-ray crystallography, a state-of-the art technique that allows scientists to see the atomic architecture of a molecule. The project represents the first time the technique has been applied to an ancient protein.
The structures allowed the scientists to identify exactly how the new function evolved. They found that just seven historical mutations, when introduced into the ancestral receptor gene in the lab, recapitulated the evolution of GR's present-day response to cortisol. They were even able to deduce the order in which these changes occurred, because some mutations caused the protein to lose its function entirely if other "permissive" changes, which otherwise had a negligible effect on the protein, were not in place first.
The most radical evolution remodeled a whole section of the protein, bringing a group of atoms close to the hormone. A second mutation in this repositioned region then created a tight new interaction with cortisol. Other earlier mutations buttressed particular parts of the protein so they could tolerate this eventual remodeling.
Contact: Joseph W. Thornton, joet@uoregon.edu; Eric Ortlund, eric.ortlund@emory.edu
See: ScienceExpress, August 16 2007; 10.1126/science.1142819 (Science Express Reports) ; Eric A. Ortlund, Jamie T. Bridgham, Matthew R. Redinbo, and Joseph W. Thornton; "Crystal Structure of an Ancient Protein: Evolution by Conformational Epistasis."
The work was funded by multiple grants from the National Institutes of Health and the National Science Foundation, the UNC Lineberger Comprehensive Cancer Center and an Alfred P. Sloan Research Fellowship to Thornton.
Original press releases can be found at: http://www.unc.edu/news/archives/aug07/grstructure081607.html
and http://waddle.uoregon.edu/?id=815
Argonne National Laboratory, the Advanced Photon Source, Stanford Linear Accelerator Center, and the Stanford Synchrotron Radiation Laboratory are funded by the U.S. Department of Energy's Office of Science.
Argonne is a U.S. Department of Energy laboratory managed by UChicago Argonne, LLC