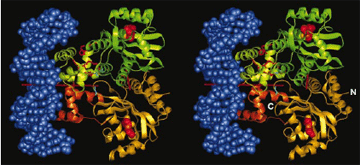
A method that cells use to communicate and coordinate activities has been confirmed by researchers from Cornell University, Monsanto Co., and Argonne. This work could lead to new drugs for the fight against such diseases as cystic fibrosis and the bubonic plague, or to new technologies that perform useful environmental tasks such as filtering water.
Biologists have theorized that bacteria communicate by releasing and sensing chemical pheromones to detect their population densities, an activity called "quorum sensing." This theory was confirmed by experimentation using Argonne's Structural Biology Center (SBC) beamlines at the Advanced Photon Source. The experimenters determined the molecular structure of a key protein - TraR - in this interbacterial communication chain and witnessed how TraR acts as a relay to sense pheromones and then activate genes to create biofilms such as the scum found on ponds.
This discovery could be the first step toward human intervention in these quorum sensing pathways, either by thwarting activities of harmful bacteria or facilitating activities of helpful ones.
Researchers at the Cornell College of Agriculture and Life Sciences isolated and purified the TraR protein and sent it to SBC crystallographers. The protein was crystallized and the crystal exposed to the high-brilliance APS x-ray beam. Computations then transformed millions of x-ray data points from the crystallized protein into an atom-by-atom, three-dimensional map of the molecule.
The result was a microsecond snapshot of quorum sensing in progress - the structure of the A. tumefaciens TraR protein, in complex with its pheromone N-3-oxooctanoyl-L-homoserine lactone and the protein-binding site on bacterial DNA.
The techniques developed by the Cornell-Argonne team and used to purify the A. tumefaciens protein and solve its structure with x-ray crystallography holds promise for the determination of protein structures in other bacteria and the potential design of specific treatments to interrupt or enhance the quorum-sensing process.
R.-g. Zhang,[1], T. Pappas[2], J. L. Brace[1], P. C. Miller[3]*, T. Oulmassov[3], J. M. Molyneaux[3]*, J. C. Anderson[3], J. K. Bashkin[3]*, S. C. Winans[2], and A. Joachimiak[1]
[1] Bioscience Division/Structural Biology Center, Argonne National Laboratory
[2] Department of Microbiology, Cornell University
[3] Monsanto Company
*Present address: Pharmacia Corporation, St. Louis, Missouri, USA For correspondence, e-mail: andrzejj@anl.gov
This work was supported by Monsanto Company; the U.S. Department of Energy (DOE), Office of Biological and Environmental Research; and a National Research Service Award to S.C.W. Argonne National Laboratory is operated by The University of Chicago for the U.S. DOE under Contract W-31-109-ENG-38. The Advanced Photon Source is funded by the U.S. DOE, Office of Science, Office of Basic Energy Sciences.
See: "Structure of a bacterial quorumsensing transcription factor complexed with pheromone and DNA," R.-g. Zhang et al., Nature 417 27, 971-974 (2002).